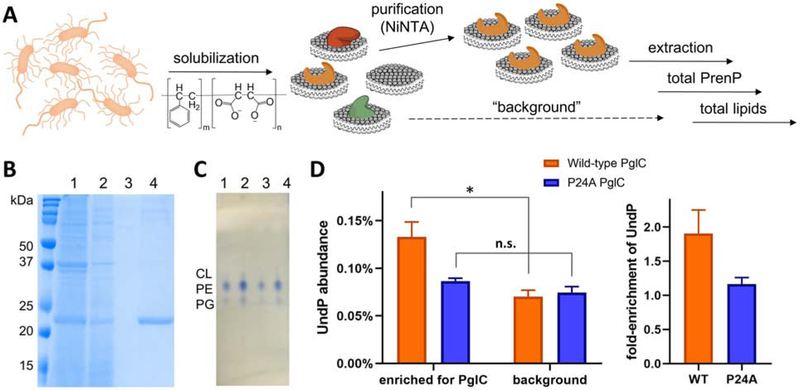
Fig. 4 – Quantification of PrenP enrichment around SMA-solubilized PglC.
A. Schematic showing work-flow for solubilization and purification of SMA-solubilized PglC-His6 and extraction and quantification of the surrounding lipids.
B. Coomassie-stained SDS-PAGE gel showing purification of SMA-lipoparticles with PglC-His6, purified by Ni-NTA affinity chromatography. Lanes: 1 = load, 2 = flow-through, 3 = wash, 4 = elution.
C. Thin-layer chromatography analysis of extracted lipids. Lanes: 1, 2 = E. coli lipid commercial standard, 5 nmol and 10 nmol respectively. Lanes 3, 4 = extracted lipids from SMA-lipoparticles, ~4 nmol and ~8 nmol respectively. CL = cardiolipin, PE = phosphotidylethanolamine, PG = phosphotidylglycerol.
D. Left: Quantification of UndP in lipid extracts from PglC- and background SMA-lipoparticles. * p = 0.02; n.s. = not significant. Right: Fold-enrichment of UndP, calculated as the UndP abundance in PglC SMA-lipoparticles over UndP abundance in background SMA-lipoparticles. Error bars are given for mean ± SD, n = 3.