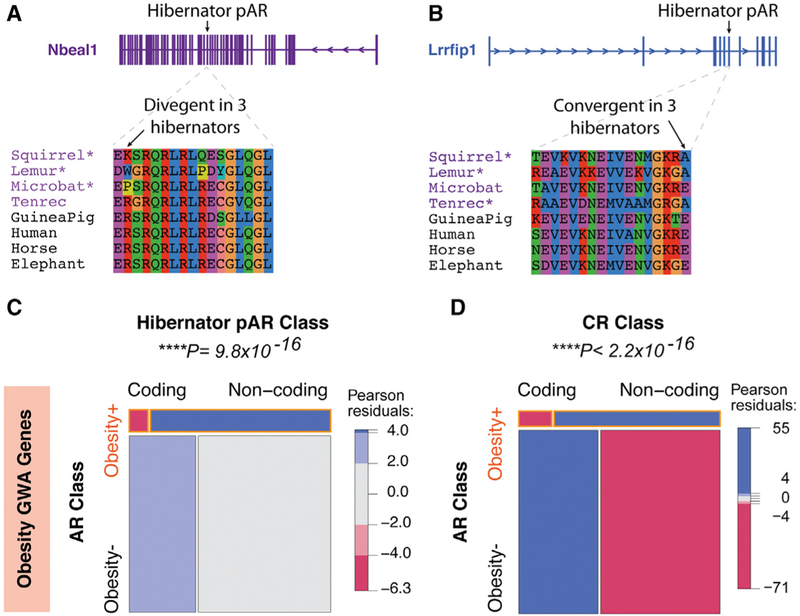
Figure 7. Parallel Coding Changes in Hibernators Are Relatively Rare in Obesity GWA Genes Compared to Changes to Noncoding cis Elements.
(A and B) Examples of parallel coding changes to amino acid sequences in hibernator pARs are shown. The two classes of parallel changes detected are divergent changes (A; Nbeal1), which involve changing the same non-hibernator conserved amino acid to different residues, and convergent changes (B; Lrrfip1), which involve changes to the same residue. Hibernators are highlighted by purple text, and non-hibernators from different clades are in black text. The exon harboring the hibernator pAR is indicated above the gene models. Hibernators with the parallel coding change are indicated by asterisks.
(C) The mosaic plot shows noncoding hibernator pARs are significantly enriched near human obesity GWA genes compared to coding pARs. Each box is proportional to the contingency count table of obesity GWA genes (obesity+) and other genes (obesity–), and subsets of obesity+ and obesity– genes linked to coding versus noncoding hibernator pARs. The chi-square test is significant (p values shown above plot), indicating that hibernator pAR associations with obesity+ genes depend significantly on whether the pARs are noncoding or coding. The mosaic plot also shows the chi-square Pearson residuals, indicating the positive (blue) and negative (red) associations in the data, and reveal that noncoding hibernator pARs are positively associated with obesity+ genes relative to coding pARs.
(D) The mosaic plot shows noncoding non-hibernator CRs are significantly enriched near human obesity GWA genes compared to CRs in coding sequence.