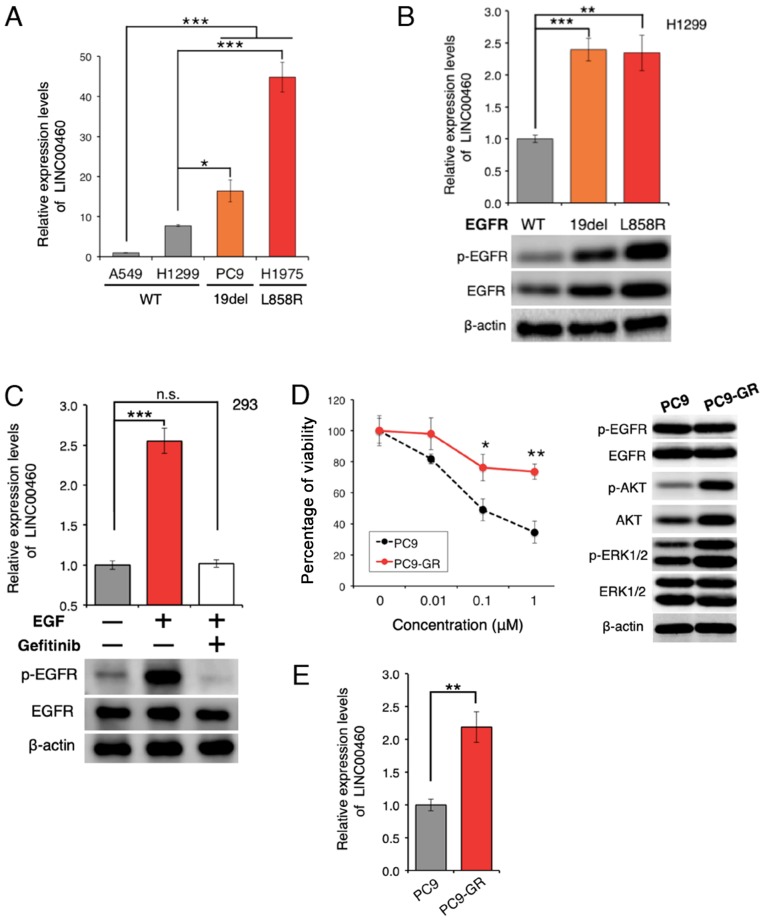
Figure 2.
Overexpression of LINC00460 in EGFR-mutant lung adenocarcinoma. (A) Relative expression of LINC00460 transcripts in cancer cell lines derived from NSCLC. LINC00460 expression was higher in cell lines harboring EGFR exon 19 deletions and L858R (PC9 and H1975) than in those without EGFR mutations (A549 and H1299) (P<0.001). The data were obtained by RT-qPCR. GAPDH mRNA was used as an internal control. (B) Upregulation of LINC00460 expression triggered by EGFR-activating mutations. LINC00460 expression was significantly upregulated in EGFR-activated mutant-positive cells. H1299 (EGFR wild-type) cells were transfected with pIRES-puro: EGFR-WT, exon 19 deletion, and L858R and selected by puromycin. (C) When EGF (200 ng/ml) was used as an EGFR activator, EGFR activation induced LINC00460 expression. EGFR inactivation was induced by pretreatment with the EGFR-TKI gefitinib (1 µM). (D) Establishment of an acquired gefitinib-resistant cell line. PC9 cells were highly sensitive to gefitinib. PC9-GR refers to gefitinib-resistant PC9 cells. PC9 or PC9-GR cells were treated with gefitinib and cell viability was determined after 72 h by WST-8 assay (left). Western blot analysis was used to observe changes in activation or expression levels of proteins in the indicated cells (right). (E) Aberrant expression of LINC00460 in gefitinib-resistant cells. LINC00460 expression levels were higher in cells with acquired resistance to gefitinib. Expression levels in PC9 and PC9-GR cells were measured by RT-qPCR. GAPDH was used as an internal control. The results of RT-qPCR are presented as the means ± SD of at least 3 independent experiments. The level of statistical significance was set at P-value <0.05 (*P<0.05, **P<0.01, ***P<0.001). n.s., not significant.