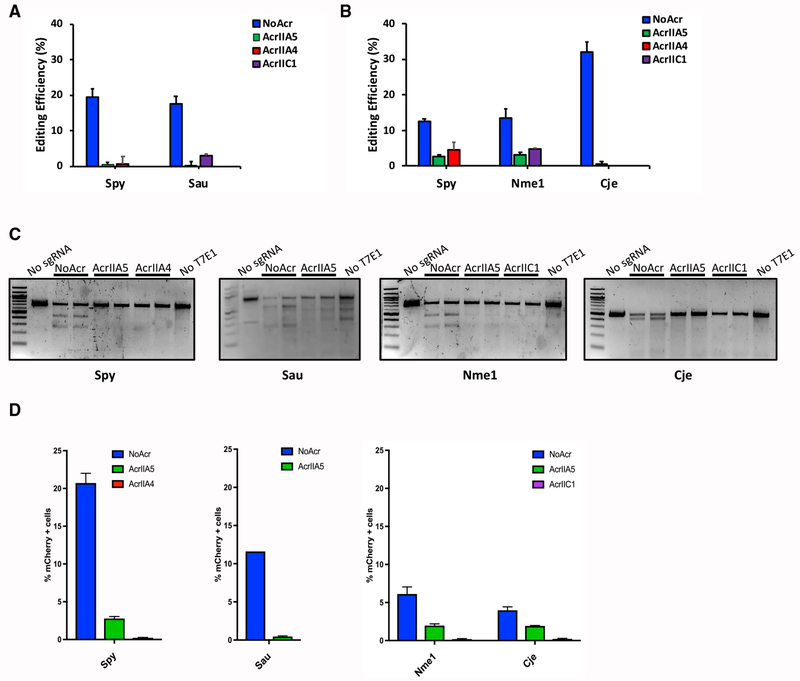
Figure 2. AcrIIA5 Inhibits Genome Editing Mediated by Nme1Cas9, SpyCas9, CjeCas9, and SauCas9 in Mammalian Cells.
(A) Co-transfection of Cas9 orthologs and their respective sgRNA expression plasmids show inhibition of genome editing by AcrIIA5 in mammalian cells. AcrIICI and AcrIIA4 are used as positive controls for SauCas9 and SpyCas9 inhibition, respectively. Genome editing was quantified using TIDE analysis (Brinkman et al., 2014) in mouse cells.
(B) Co-transfection of Cas9 orthologs and their respective sgRNA expression plasmids targeting either the ARHGEF9 (SpyCas9 and Nme1Cas9) or AAVS1 locus (CjeCas9) show inhibition of genome editing by AcrIIA5 in a human HEK293T cell line. Editing was quantified by TIDE analysis (Brinkman et al., 2014). AcrIIC1 and ArIIA4 were used as positive controls for type II-C (Nme1Cas9 and CjeCas9) and type II-A (SpyCas9) inhibition, respectively.
(C) Genome editing in the cell lines used in (A) and (B) were analyzed by T7E1 experiments. The image is representative of at least three replicates.
(D) Co-transfection of Cas9 orthologs, their respective sgRNAs, and AcrIIA5 expression plasmids in a cell line stably expressing TLR-MCV1, a variation of the traffic light reporter (TLR) system (Certo et al., 2011). TLR-MCV1 contains an artificial locus harboring target sites for a wide range of Cas9 orthologs. Upon double-strand break induction by a Cas9 ortholog, a subset of non-homologous end-joining (NHEJ) repair events generate indels that place mCherry in frame. The percentage (%) of mCherry cells was used as an estimate of genome-editing efficiency. Anti-CRISPRs used as controls were AcrIIC1 for type II-C Cas9 homologs (Nme1Cas9 and CjeCas9) and AcrIIA4 for type II-A SpyCas9.
In (A), (B), and (D), the values and error bars represent the mean ± the SD of three independent biological replicates.