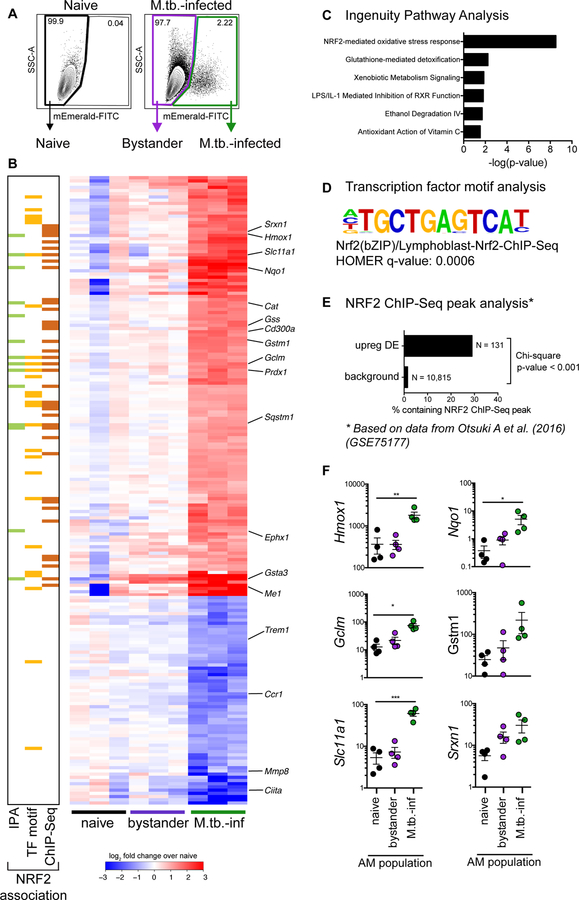
Figure 2: M.tb.-infected alveolar macrophages up-regulate an NRF2-associated antioxidant gene signature.
(A) Gating scheme to sort naïve, bystander, and M.tb.-infected AMs from bronchoalveolar lavage (BAL) samples after high dose aerosol infection with mEmerald-tagged H37Rv. (B) Heatmap of gene expression (log2 fold change over average of naïve AMs) for 196 differentially expressed genes between naïve and M.tb.-infected AMs (Filtering criteria: average CPM >1, |fold change| > 2 and FDR < 0.01, Benjamini-Hochberg calculated). Columns are independent experiments (pooled mice) and rows are genes. Genes called out are known NRF2 target genes of interest as well as downregulated pro-inflammatory genes. Colored bars to the left indicate NRF2 association as determined by 3 different methods: (C) Ingenuity Pathway Analysis, (D) transcription factor promoter binding motif enrichment analysis (HOMER), and (E) ChIP-seq peak analysis. (F) RT-qPCR validation of NRF2 associated genes for naïve, bystander and M.tb.-infected AMs 24 hours post-infection. Values are relative to Ef1a. Data is presented from 3 independent experiments with one-way ANOVA with Dunnett’s post-test *p < 0.05, **p < 0.01, ***p < 0.001.