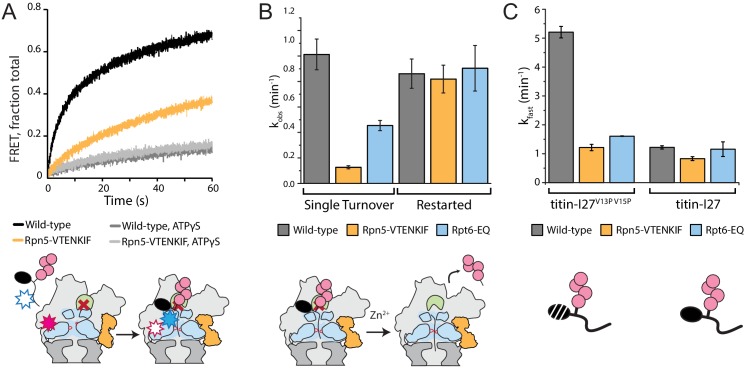
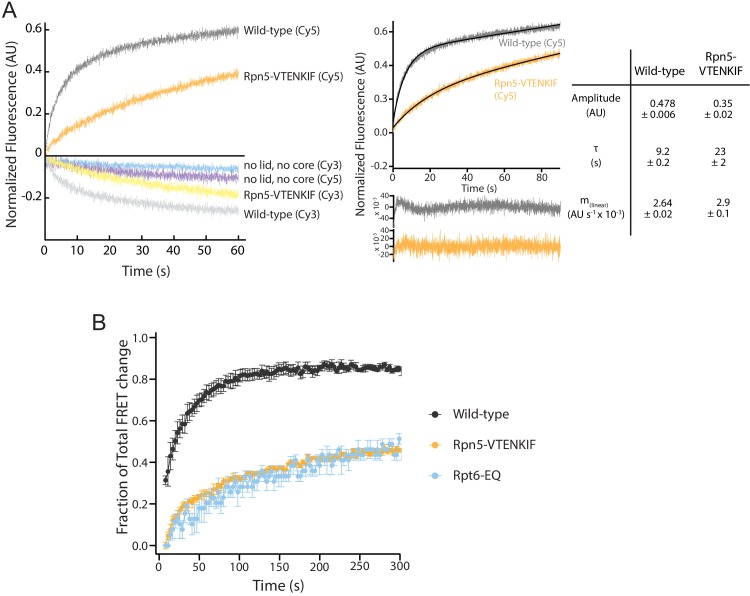
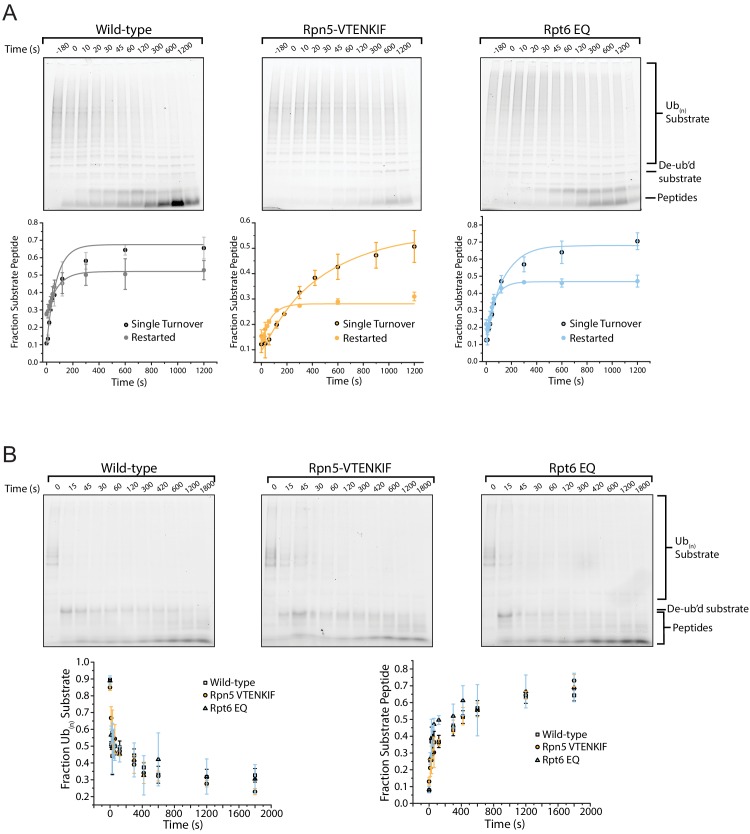
Figure 5. Disrupting the conformational equilibrium inhibits substrate tail insertion but not later steps of degradation.
(A) Representative traces for the increase in acceptor fluorescence/FRET upon insertion of the ubiquitinated FAM-titin-I27V15P-Cy5 substrate’s flexible initiation into the central pore of wild-type and Rpn5-VTENKIF mutant proteasomes with o-PA inhibited Rpn11, in the presence of ATP or ATPγS. The schematic below depicts the experimental setup, where FRET occurs when a substrate’s flexible initiation region labeled with an acceptor dye (blue star) enters and then stalls in the central pore of a proteasome containing inhibited Rpn11 (red cross) and a donor dye (red star) near the processing channel. The substrate’s ubiquitin modification is represented in pink, the Rpt ring is shown in light blue, the core particle in dark grey, and Rpn5 in orange. (B) Rate constants for the single-turnover, ubiquitin-dependent degradation of ubiquitinated G3P model substrate, either without stalling the proteasome (left) or after stalling translocation for 3 min with o-PA inhibited Rpn11 and restarting by the addition of Zn2+ (right). Rates were determined from single-exponential fits of the appearance of fluorescently tagged peptide products on SDS PAGE gels. Error bars represent SEM for the fit, N ≥ 3, technical replicates. (C) Ubiquitin-dependent degradation rates for wild-type, Rpn5-VTENKIF and Rpt6-EQ mutant proteasomes degrading the destabilized FAM-titin-I27V13P/V15P-35mer tail or the non-destabilized FAM-titin-I27-35mer tail substrate under single-turnover conditions. Shown are the rate constants for the dominant fast phase derived from a double-exponential fit of the degradation kinetics (N = 3, technical replicates, error bars represent SD).