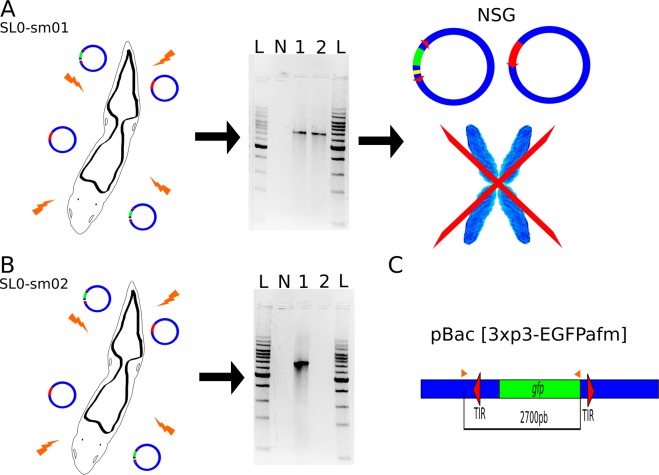
Figure 2.
Experimental scheme testing the integration of the gfp gene into the Stenostomum genome. (A) SL0-sm01 electroporated worms using pBac [3xp3-EGFPafm] and pB∆Sac plasmids were tested by PCR (gel lanes: L = Kb ladder; N = control/ no DNA; 1 = worm sample three days after electroporation; 2 = worm sample 15 days after electroporation). Amplicons of 2,700 bp indicated that precise transposition had not occurred (see scheme C). Illumina NSG using gDNA isolated six months after electroporation allowed assembly of both plasmids (complete and circularized). However, no indication of integration into the worm’s chromosomes was obtained. (B) SL0-sm02 electroporated worms using pBac[3xp3-EGFPafm] and pB∆Sac plasmids tested by PCR 3 and 15 days after electroporation. No amplicons were obtained 15 days after electroporation, showing the plasmid was lost in this period; (C) scheme of the pBac[3xp3-EGFPafm] plasmid containing piggyBac TIRs sequences (red arrowhead) and gfp gene (green), other plasmid sequences (blue) and primers used for PCRs (orange arrows). Amplicons containing 81 bp of plasmid sequences upstream of the TIRs, suggest that a precise transposition did not occur.