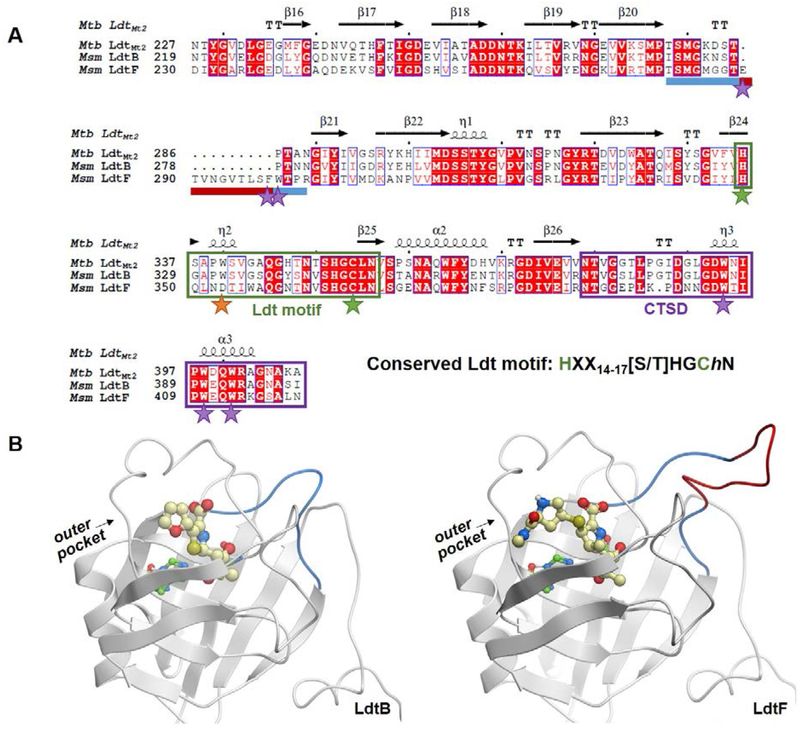
Figure 2.
Comparison of mycobacterial class 2 and class 6 Ldt catalytic domains. A) A sequence alignment of Msm LdtB and LdtF and Mtb LdtMt2. The LdtF 10-residue insertion is indicated by a maroon bar and the rest of the loop between β-strands 20 and 21 is indicated by a blue bar. The Ldt motif is boxed in green, and catalytic cysteine and histidine residues are noted with green stars. The conserved tryptophan residue (an aspartate in LdtF) is noted with an orange star, and additional residues discussed are highlighted with purple stars. The CTSD is boxed in purple. The alignment was performed using Molsoft ICM-Chemist-Pro26 and visualized with ESPript 3.0.27 B) Homology models of LdtB-faropenem (left) and LdtF-meropenem (right) indicate the location of the LdtF insertion (red). The LdtF insertion is near the active site, as indicated by the modeled (carba)penems. Faropenem and meropenem are shown as yellow balls and sticks.