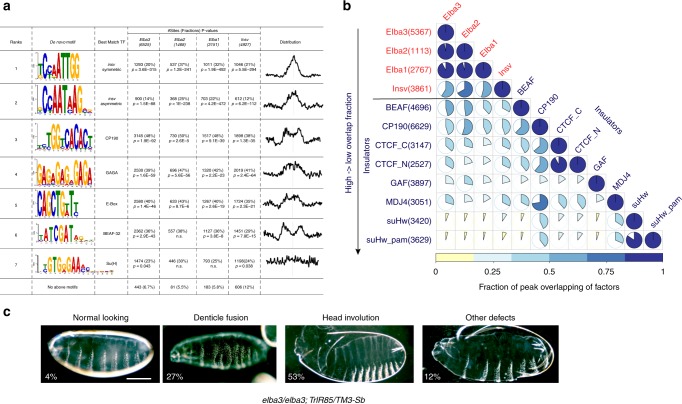
Fig. 5. Interaction of ELBA and Insv with other insulator proteins.
a The de novo motif discovery analysis from the Elba and Insv peaks identified the Insv/ELBA symmetric and asymmetric motifs, motifs for CP190, GAF, and BEAF-32, E-box and the Su(H)-binding site. b The pairwise peak overlapping matrix summarizes the genomic co-occupancy of the ELBA factors, Insv and six known insulator proteins (see Methods). Among these insulator proteins, CP190 exhibits the highest overlap with ELBA and Insv, followed by BEAF-32 and CTCF. ELBA and Insv have the least overlapping with GAF, Mod(Mdg4), and Su(Hw). c Representative images of cuticle preps from the mutants for the ELBA factors and Trl (GAF) or CP190. The percentages are shown for animals with the elba3 homozygous with the TrlR85 heterozygous mutations. Scale bar: 100 µm. Source data of phenotypic quantification is available in a Source Data file.