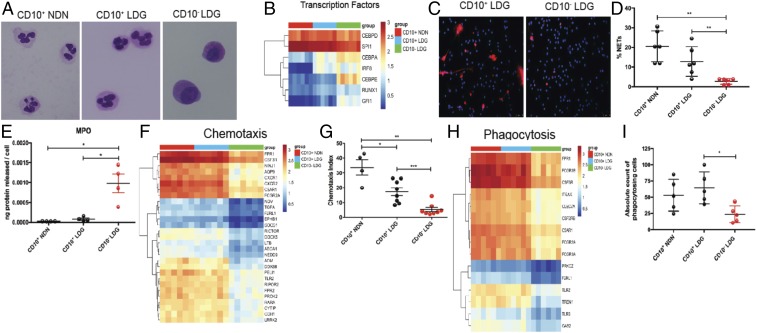
Fig. 2.
Lupus neutrophil subsets differ phenotypically and functionally. (A) Representative images of Giemsa-stained sorted lupus CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 4); original magnification 60×. (B) RNA-seq analysis of CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 6/group) for TFs involved in myeloid development. (C and D) Representative images of cells undergoing NET formation in unstimulated sorted CD10+ LDGs and CD10− LDGs after a 2-h incubation (n = 5/group). MPO is in red and DNA is in blue; original magnification 40×. (E) MPO ELISA of culture supernatants of unstimulated CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 4/group) after a 2-h incubation. (F) RNA-seq analysis for genes involved in chemotaxis in lupus neutrophil subsets. (G) fMLP-induced chemotactic index in lupus neutrophil subsets after a 2-h incubation (n = 4 for CD10+ NDNs; n = 8 for CD10+ and CD10− LDGs). (H) Expression of phagocytosis genes by RNA-seq analysis. (I) Phagocytosis of S. aureus bioparticles for lupus neutrophil subsets (n = 5). Results for all measurements are mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.