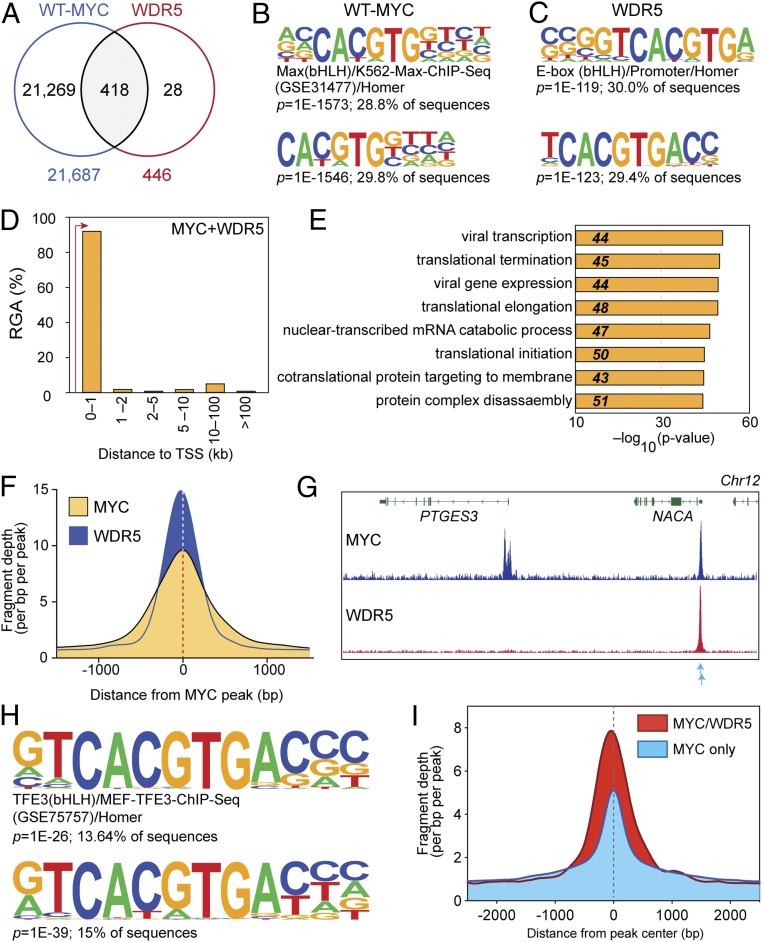
Fig. 2.
MYC and WDR5 colocalize at cohort of protein synthesis genes in Ramos cells. (A) Venn diagram, showing overlap of HA–WT–MYC and WDR5-binding sites—as determined by ChIP-Seq (false discovery rate [FDR] = 0.01). HA–WT–MYC was monitored in switched WT cells; WDR5 was monitored in unswitched WBM cells that express endogenous, wild-type, MYC protein. (B) Motif analysis for HA-tagged MYC-binding sites. At Top is known-motif analysis; Bottom is de novo motif analysis. Significance values and the percentage of ChIP-Seq peaks containing the indicated motif are shown for each. (C) As in B, except for WDR5-binding sites. (D) Distribution of shared MYC- and WDR5-binding sites, binned according to distance from the nearest annotated TSS. RGA, region–gene association. (E) The top 8 GO enrichment categories for genes bound by MYC and WDR5 in Ramos cells. (F) Histogram of normalized MYC and WDR5 ChIP-Seq fragment counts at MYC–WDR5 cobound sites in a 1,500-bp window either side of the center of the MYC peaks. (G) IGV screenshot of representative ChIP-Seq data for HA–WT–MYC and WDR5 in Ramos cells. Shown is the PTGES3 gene, which is bound only by MYC, and NACA, which is bound by both MYC and WDR5. Blue arrows indicate the position of 2 imperfect E-boxes in the NACA locus. n = 3 independent ChIP-Seq experiments for MYC and WDR5. (H) Motif analysis for MYC-binding sites at WDR5 cobound genes. At Top is known-motif analysis; Bottom is de novo motif analysis. Other details are as in B. (I) Histogram of normalized MYC ChIP-Seq fragment counts at sites with imperfect E-boxes that are bound by both MYC and WDR5 (red) or MYC only (blue). A 2,500-bp window either side of the peak center is shown.