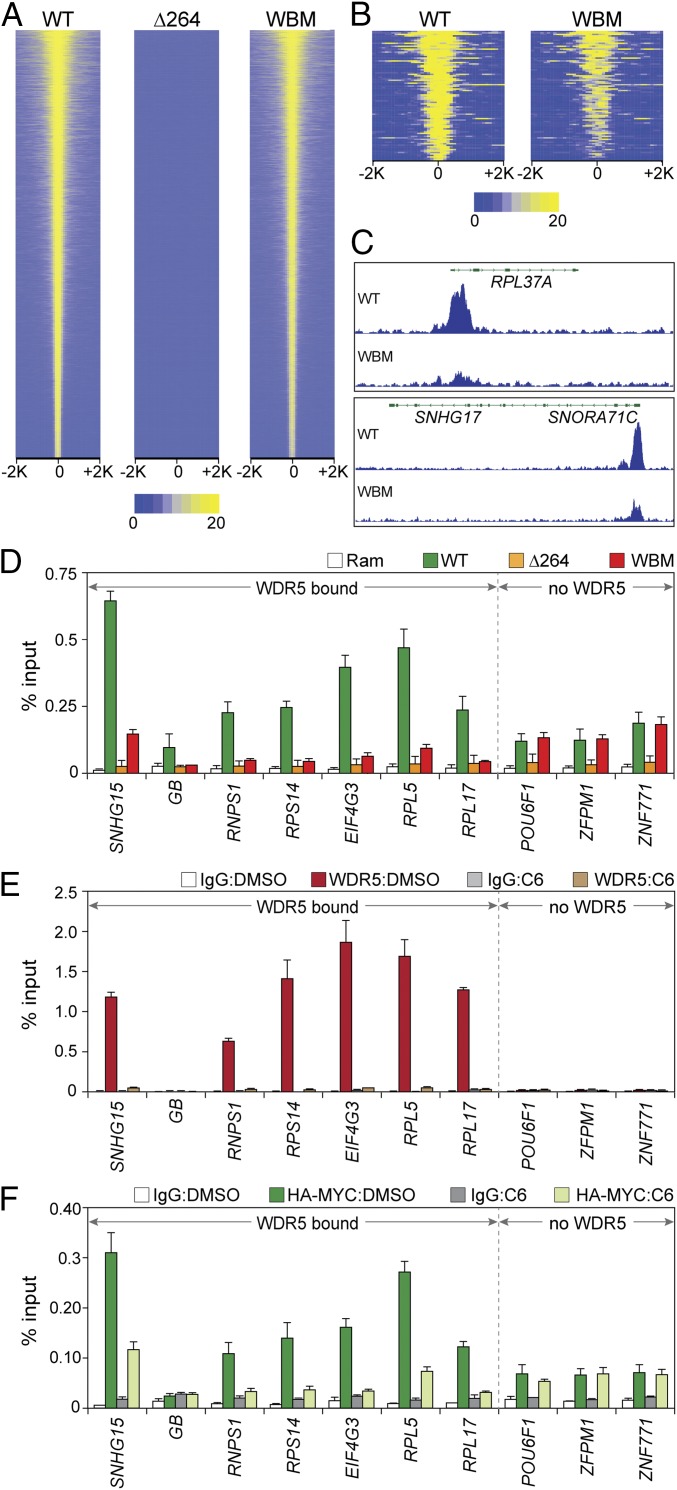
Fig. 3.
Interaction of MYC with protein synthesis genes is WDR5 dependent. (A) Heatmap, showing HA-tagged MYC peak intensity for each MYC protein (ChIP-Seq) after switching; the figure displays the combined average of normalized peak intensity (read density) in 100-bp bins ±2 kb around peak centers. Intensities are ranked based on WT–MYC. (B) As in A, except for the 88 loci where the WBM mutant was associated with a significant reduction in HA–MYC binding. (C) Two IGV screenshots of normalized representative ChIP-Seq data for HA-tagged MYC (WT or WBM) in the indicated cell lines after switching. Both RPL37 and SNROA71C/SNHG17 bind WDR5. n = 3 independent ChIP-Seq experiments for HA-tagged MYC proteins. (D) ChIP was performed (anti-HA antibody) in cells switched to the indicated MYC proteins (or parental Ramos cells). Coprecipitating DNAs were monitored (Q-PCR) with primer sets spanning ChIP-Seq peaks. Six WDR5-bound loci, and 3 non–WDR5-bound loci, were probed, as indicated. GB, corresponds to the SNHG15 gene body, where little MYC is detected. ChIP signal is represented as a percentage of input DNA. (E) WT engineered Ramos cells were selected after switching to permanently express HA-tagged WT MYC. These cells were then treated with either DMSO or 25 µM C6 for 4 h and WDR5 binding to the indicated loci probed via ChIP-qPCR. IgG is a negative control ChIP. Primer sets and abbreviations are as in D. (F) As in E, except that ChIP-qPCR was performed for MYC. The mean and SD of 3 experiments are shown.