Figure 1.
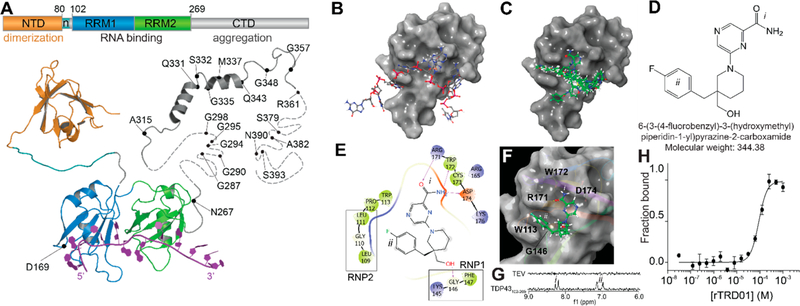
In silico docking to the RNA recognition motif of TDP-43 identifies compound rTRD01. (A) Composite linear and structure representation of TDP-43. The N-terminal domain (NTD) of TDP-43 (orange) is important for dimerization and also contains a nuclear localization signal, n (cyan) (PDB code 2n4p25). The RNA recognition motifs 1 and 2 (RRM1 and 2, blue and green, respectively, PDB code 4bs217) interact with RNA (purple). The C-terminal domain (CTD, gray) is largely unstructured and is considered a “prion-like” domain. Residues M311–Q360 from the CTD were resolved using NMR (PDB 2n3x41). In unstructured regions, each dash represents a residue. ALS-associated mutations are noted. (B) TDP-43 RRM1 domain (surface representation) in complex with DNA (stick and ball representation) (PDB code 4iuf13). (C) Top 10 compounds (green, sticks and balls representation) from in silico docking on TDP-43 RRM1. (D) Structure of rTRD01 (6-(3-(4-fluorobenzyl)-3-(hydroxymethyl)piperidin-1-yl)pyrazine-2-carboxamide), where i is a pyrazinamide group and ii is a fluorobenzyl group shown to interact with TDP-43 by STD NMR (8.2 ppm and 6.9–7.2 ppm, respectively). (E) 2D representation of rTRD01 binding pocket, which includes residues from the ribonucleoprotein motif 1 (RNP1) and ribonucleoprotein motif 2 (RNP2) of TDP-43 RRM1. (F) Docking pose of rTRD01 (green, sticks and balls representation) on RRM1 domain of TDP-43 (surface representation). (G) 1D 1H STD NMR showing on-resonance difference spectrum of 500 μM rTRD01 with 5 μM TDP-43102–269. The aromatic region of the NMR spectrum (6–9 ppm) is shown. (H) MST values from thermographs of NT-647 labeled TDP-43102–269 in the presence of increasing concentrations (0.03 μM to 1 mM) of rTRD01were used to determine dissociation constant for binding of rTRD01 to TDP-43102–269. The data were fitted as described in the Methods section yielding an apparent Kd of 89.4 ± 0.8 μM (r2 = 0.93). Data are given as mean ± SEM (n = 3 independent replicates).