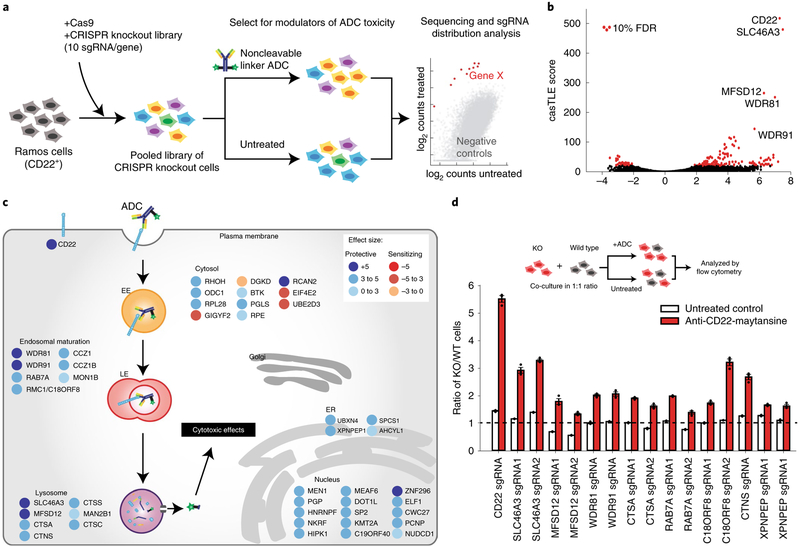
Fig. 1 |. Genome-wide CRISPR screen uncovers diverse endolysosomal regulators of ADC toxicity.
a, Schematic for the genome-wide screen. Ramos cells expressing Cas9 were infected with a lentiviral, genome-wide sgRNA library, the population was split and half the cells were treated with anti-CD22-maytansine ADC for three rounds, killing ~50% cells each round. The resulting populations were subjected to deep sequencing and analysis. The screen was performed in duplicate. b, Volcano plot of all genes indicating effect and confidence scores for genome-wide ADC screen. Effect and confidence scores determined by casTLE. c, Schematic for top 30 and selected endolysosomal trafficking regulator hits in genome-wide screen, color coded by effect size (calculated by casTLE). Subcellular localization is indicated according to gene ontology annotations. EE, early endosomes; LE, late endosomes; ER, endoplasmic reticulum. d, Validation of hits in Ramos cells using competitive-growth assays: cells expressing sgRNAs for knockout (KO) of indicated genes (mCherry+) and control (mCherry−) were cocultured in 1:1 ratio. Cells were either treated with anti-CD22-maytansine ADC or left untreated for 3 d. Resulting ratio of KO:control was determined using flow cytometry. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. WT, wild type.