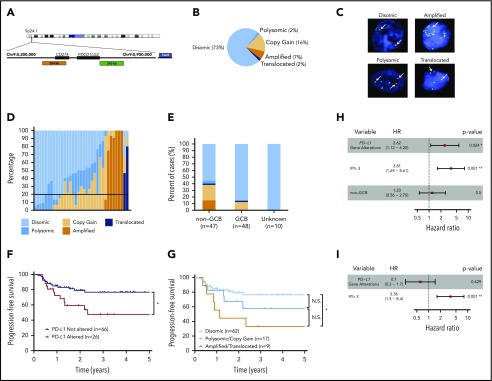
Figure 1.
PD-L1 gene alterations are recurrently observed in DLBCL and are associated with inferior PFS to front-line chemoimmunotherapy. (A) Chromosome 9 structure. Enlarged below is the 9p24.1 region encoding PD-L1 and PD-L2. PD-L1, centromere 9, and translocation FISH probes are shown in orange, blue, and green respectively. (B) Incidence of PD-L1 alterations according to underlying mechanism. (C) Representative DLBCL cells with the indicated PD-L1 locus status as assessed by FISH. Arrows indicate signals for PD-L1 (orange) and translocation (green) probes. Split signals are seen between PD-L1 and translocation probes in the representative DLBCL with a PD-L1 translocation. (D) Waterfall plot demonstrating the frequency of lymphoma cells (30 analyzed/specimen) with the indicated PD-L1 locus status. The horizontal black line represents the minimum number of lymphoma cells required for a case to be considered PD-L1 altered (20% of cells). (E) Incidence of PD-L1 alterations by cell of origin (*P < .05, Fisher’s exact test). (F) PFS of DLBCL patients with or without PD-L1 gene alterations. (*P = .024, log-rank test). (G) PFS of DLBCL patients according to specific PD-L1 alteration (*P = .012, log-rank test). Clinical and genetic univariate (H) and multivariate (I) risk models for PFS in DLBCL.