-
A
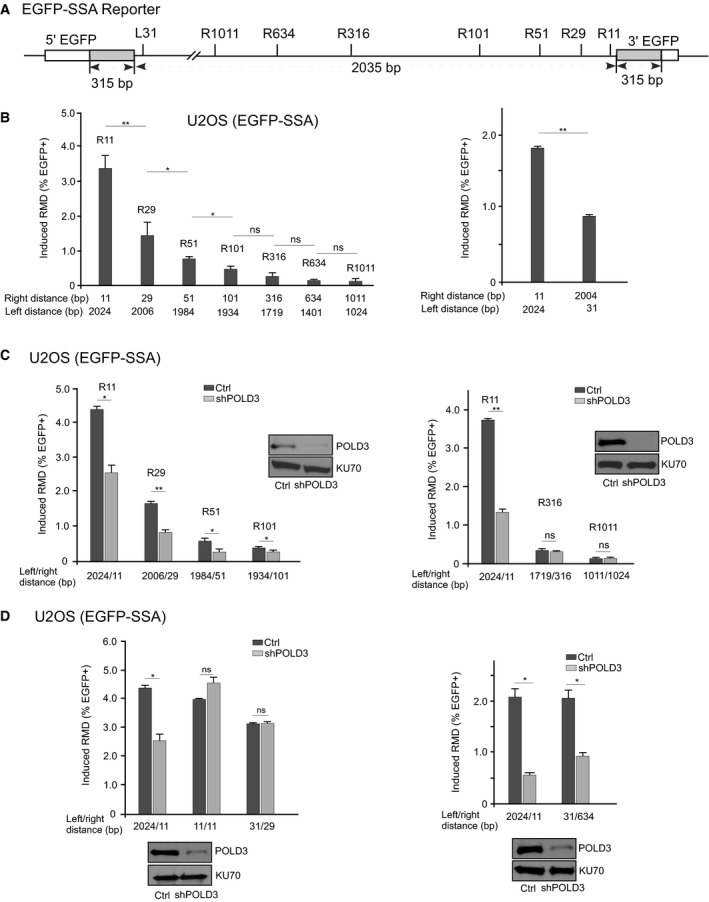
A diagram indicating the position of DSBs generated by sgRNAs/Cas9 on the EGFP‐SSA reporter.
-
B
RMD efficiency is decreased when the distance from a DSB to one repeat is increased. RMD assays were performed in U2OS (EGFP‐SSA‐3427‐17) cells using the indicated sgRNAs/Cas9. The distance between the sgRNA/Cas9 cleavage sites and the repeat sequences is shown.
-
C, D
Knockdown of POLD3 impairs RMD when a DSB is close to one repeat. RMD frequency was determined in U2OS (EGFP‐SSA‐3427‐17) cells using the indicated sgRNAs/Cas9 with or without depletion of POLD3 by lentivirus‐expressed shRNA. Cells infected with a lentiviral empty vector were used as control. The distance between the sgRNA/Cas9 cleavage sites and the repeat sequences is indicated. The knockdown level of POLD3 was shown by Western blot, and KU70 was used as the loading control.
Data information: Error bars represent standard deviation (SD) of at least three independent experiments. *
P < 0.05, **
P < 0.01, ns, not significant, two‐tailed non‐paired
t‐test.