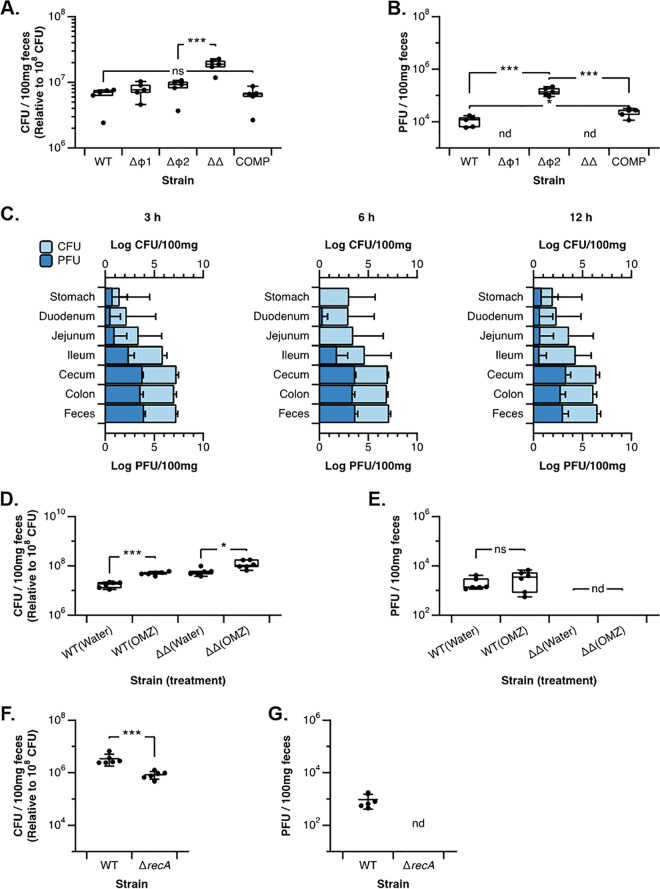
FIG 3.
Characterization of L. reuteri 6475 prophages during gastrointestinal transit. (A) Cell numbers of the L. reuteri wild-type strain (WT), the L. reuteri ΔΦ1, ΔΦ2, and ΔΦ1 ΔΦ2 (ΔΔ) mutant strains, and the L. reuteriΔΦ1 ΔΦ2∷Φ1∷Φ2 complemented strain (COMP) as determined by bacterial culture from feces. ns, not significant; ***, P < 0.001. (B) Phage numbers expressed as PFU derived from mice administered the WT, ΔΦ1, ΔΦ2, ΔΔ, and COMP strains. n = 5 animals per group. nd, not detected; ***, P < 0.001. (C) Temporal and spatial analyses of the cell numbers of L. reuteri 6475 wild-type (light blue bars, top axis) and PFU (dark blue bars, bottom axis) normalized to 100 mg tissue/feces following administration of 108 CFU at t = 0 h. Per time point, five animals were sacrificed. (D and E) Bacterial (D) and phage (E) counts following administration of L. reuteri strain 6475 (WT) or the ΔΦ1 ΔΦ2 mutant (ΔΔ) to mice that received regular drinking water (Water) or drinking water supplemented with omeprazole (OMZ). n = 6 animals per treatment group. (F and G) Bacterial (F) and phage (G) counts following administration of L. reuteri 6475 (WT) or 6475 ΔrecA (ΔrecA). For all panels, CFU data are expressed per 100 mg feces and normalized to 108 administered CFU and PFU data are expressed per 100 mg feces. ns, not significant; nd, not detected. *, P < 0.05; ***, P < 0.001 (t test analyses).