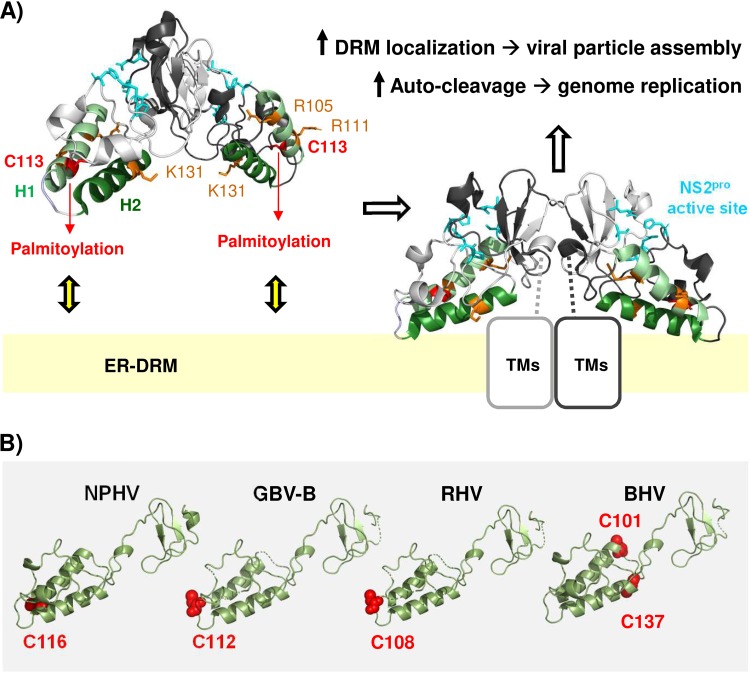
FIG 8.
Model for the functional role of NS2/C113 palmitoylation. (A) NS2/C113 palmitoylation in helix 1 (H1), potentially assisted by positively charged residues in H1 and H2, promotes the membrane association of H1 and H2, properly orienting the composite catalytic domains of dimeric NS2 toward the cytoplasm for efficient autocleavage at the NS2-NS3 junction site and facilitating the association of N-terminal transmembrane domains of NS2 (TMs) with the ER-DRM, thereby enhancing HCV particle assembly (see Discussion for details). The structural model was based on the crystal structure of the gt1a HCV NS2 protease domain (RCSB Protein Data Bank [PDB] entry 2HD0) (11; http://www.rcsb.org). (B) We used the PyMOL Molecular Graphics System v2.0.6 (DeLano Scientific, San Carlos, CA, USA) (http://www.pymol.org) to create the protein structure images. The protein sequences were retrieved from the NCBI GenBank database (51): NPHV H3-011 (JQ434008), GBV-B (AY243572), RHV NLR07-oct70 (KC411784), and BHV PDB-452 (KC796090). An alignment of sequences was computed by MUSCLE (52) and improved by minor manual modifications in SeaView (53) according to the alignment published by Boukadida et al. (12). Homology-derived three-dimensional (3D) structure models for New World primate hepacivirus (GBV-B) and equine (NPHV), rodent (RHV), and bat hepaciviruses (BHV) were constructed using the WHAT IF modeling server (54) with sequence structure alignments based on PDB entry 2HD0 (chain A) from the RCSB PDB (http://www.rcsb.org) (11), comprising the crystal structure of the catalytic domain of the hepatitis C virus NS2-3 protease from genotype 1a (isolate H).