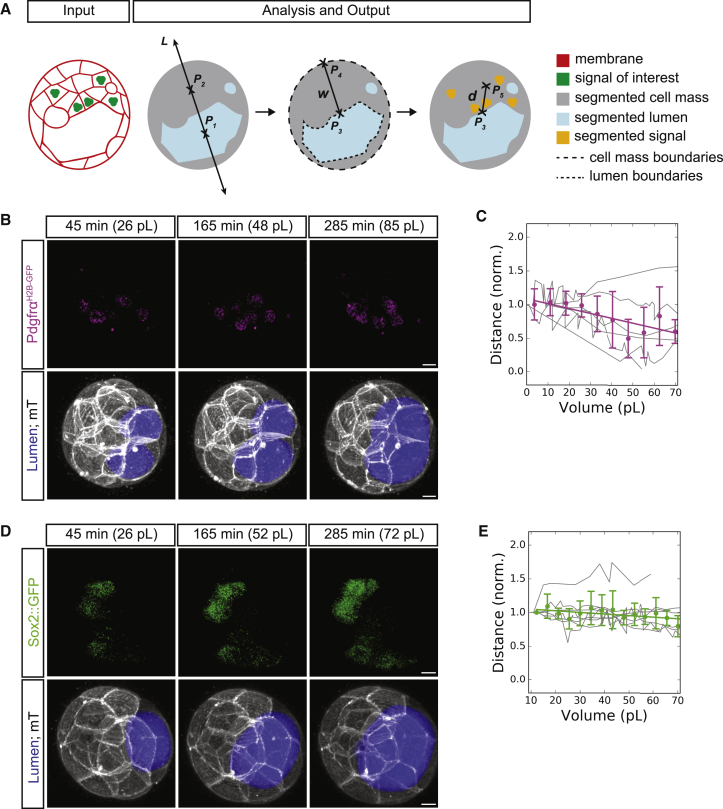
Figure 3.
The Pdgfr⍺ Signaling Domain Approaches the Luminal Surface as the Lumen Grows in Volume
(A) Schematic 2D representation of 3D analysis method for the tracking and normalization of fate reporter expression proximity to the ICM-lumen interface. are 3D points. is a 3D line ( equivalent) that defines the embryonic-abembryonic axis. is the 3D line segment ( equivalent) that measures the ICM width. is the 3D line segment ( equivalent) that measures the distance from the center of mass of the signal of interest to the ICM-lumen interface. See Image Analysis for formal definitions of all geometric entities.
(B) Time-lapse of an E3.0 embryo expressing a PrE reporter (Pdgfr⍺H2B-GFP/+; top), a membrane marker and lumen segmentation (bottom). t = 0 min is defined as the first moment when a lumen can be segmented. Lumen volume (pL) is given for each time point shown.
(C) Quantification of the distance of the center of Pdgfr⍺ signaling domain to the surface of the lumen over time (N = 6 embryos, thin gray lines are traces of individual embryos, magenta dots are binned averages with vertical capped lines showing standard deviations, thick magenta line is the linear regression y = −0.007x + 1.090, r2 = 0.653, p < 0.005).
(D) Time-lapse of an E3.0 embryo expressing a cytoplasmic EPI reporter (Sox2::gfp; top), a membrane marker and lumen segmentation (bottom). t0 is defined as the first moment when a lumen can be segmented. Lumen volume (pL) is given for each time point shown.
(E) Quantification of the distance of the center of Sox2 expression domain to the surface of the lumen over time (N = 8 embryos, thin gray lines are traces of individual embryos, green dots are binned averages with vertical capped lines showing standard deviations, thick green line is the linear regression y = −0.002x + 1.074, r2 = 0.339, p < 0.030). All scale bars, 10 μm.