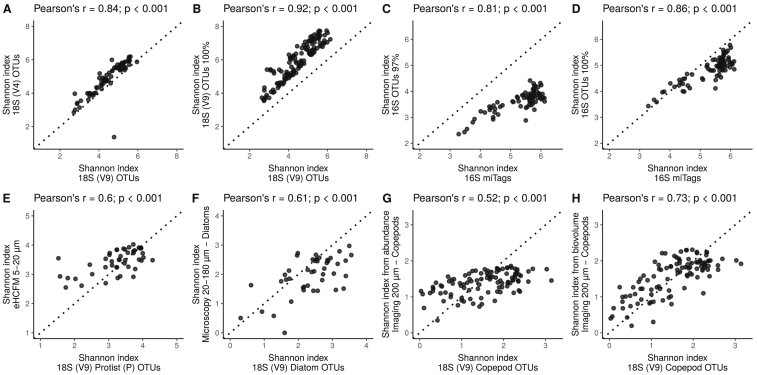
Figure S6.
Correlation between the Shannon Values Derived from Multiple Datasets of Tara Oceans, Related to STAR Methods
(A) OTUs (as defined with “swarm”; Mahé et al., 2014) obtained with the V9 (x axis) and V4 (y axis) regions of the 18S rRNA gene using surface water samples (SRF); size fraction 0.8-2000 μm; (B) OTUs either as defined with swarm (x axis) or defined at 100% sequence identity from the V9 region of the 18S rRNA gene for SRF samples, size fraction 0.8-2000 μm; (C-D) 16S rRNA gene miTags (x axis) versus OTUs defined at 97% [C] and 100% sequence similarity [D] (y axis) obtained from the V4-V5 regions of the 16S rRNA gene for SRF samples, size fraction 0.22-3 μm. (E) OTUs of photosynthetic protists obtained with the V9 region of the 18S rRNA gene (x axis) versus protists (mostly photosynthetic) as identified with environmental High Content Fluorescence Microscopy (eHCFM; data from Colin et al., 2017) in SRF-DCM samples, size fraction 5-20 μm; (F) Diatom OTUs obtained with V9 region of the 18S rRNA gene (x axis) versus diatom species counted by light microscopy (y axis) in SRF samples; size fraction 20-180 μm; (G/H) Copepod OTUs obtained with the V9 region of the 18S rRNA gene, SRF samples, size fraction 180-2000 μm (x axis) versus abundances [G] and biovolumes [H] of copepods collected by the WP2 net, > 200 μm. Inset titles show the Pearson’s correlation coefficient and its associated p value. Note the differences in axes scales. Dashed line represents 1:1 relation. Refer to STAR Methods for details on each method.