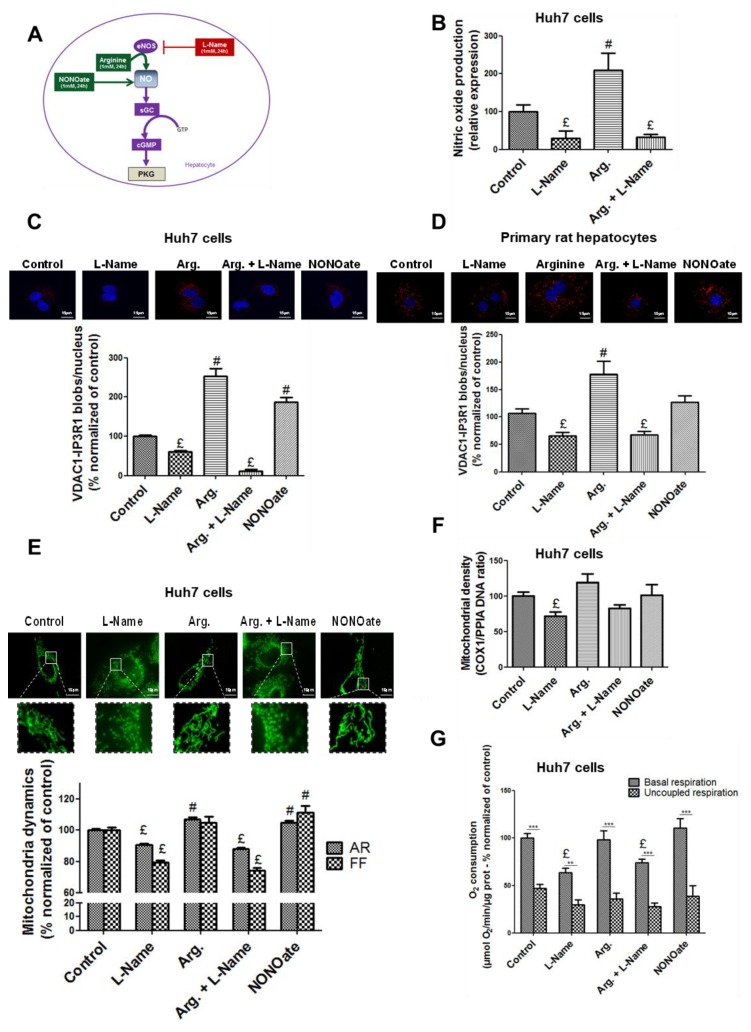
Figure 3.
NO regulates the ER–mitochondria contact points and the mitochondrial network architecture, density, and respiratory activity in hepatocytes in vitro. (A) Representative scheme of strategies used to modulate NO concentration in vitro. Arginine is a substrate of eNOS, l-Name in an inhibitor of eNOS, and NONOate (diethylamine NONOate sodium salt hydrate) is a NO donor. (B) Impact of l-Name (1 mM), arginine (1 mM), and a combination of the two on NO concentration in fresh Huh7 cells assessed using Daf-FM (15 µM). (C–G) Impact of modulating NO concentration for 24 h using l-Name (1 mM), arginine (1 mM), a combination of the two, and NONOate (1 mM), on (C,D) ER–mitochondria interactions assessed through VDAC1/IP3R1 (voltage dependent anion channel 1/inositol 1,4,5-trisphosphate receptor) interactions using in situ proximity ligation assay (PLA) in (C) Huh7 cells and (D) primary rat hepatocytes (n = 10 images minimum per experiment, three independent series per treatment, representative image at top and enlarged in Figure S7A,B, and quantitative analysis below, scale bar 15 µm, ×100); (E) mitochondrial network architecture, assessed in Huh7 cells using MitoTracker® Green (500 nM) and calculation of aspect ratio (AR) and form factor (FF) (n = 10 images minimum per experiment, three independent series per treatment, representative image at top and enlarged in Figure S7C, scale bar 15 µm, ×100); (F) mitochondrial (COX-1, cyclooxygenase-1) DNA content relative to nuclear (PPIA, peptidylprolyl isomerase A) DNA assessed in Huh7 cells using RT-qPCR (n = three independent experiments); (G) whole cell oxygen consumption assessed using oxygraphy in Huh7 cells (n = minimum six independent experiments). £ and #, p < 0.05 vs. control; ***, p < 0.05 vs. respective uncoupled state.