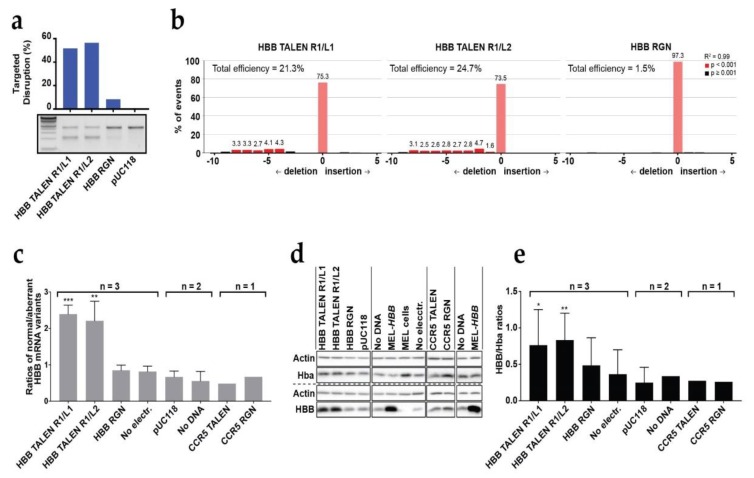
Figure 1.
DARE-based correction in humanized transgenic MEL-HBBIVS bulk cell populations. HBB-specific designer nucleases were applied in comparison to CCR5-specific designer nucleases and the GLOBE gene addition vector as a negative and positive reference, respectively, for HBB induction. (a) The T7 endonuclease I assay depicting HBBIVSI-110(G>A)-targeted disruption in TALEN- (R1/L1 or R1/L2) and RGN-edited cells relative to nuclease-free, pUC118-transfected negative controls (pUC118). (b) TIDE analysis depicting HBBIVSI-110(G>A)-targeted disruption efficiencies of TALEN- (R1/L1 or R1/L2) and RGN-edited cells relative to pUC118. Same-size indels events (-10 deletions to +10 insertions) are scored as a percentage of the total number of events. Significance cutoff was the TIDE default (p value < 0.0001). (c) The splice correction is shown at the transcript level as the ratios of normal/aberrant HBB mRNA levels (±SD) on day 3 of induced differentiation. Significant group-wise comparisons of ratios to no-electroporation controls for samples analyzed in triplicate by unmatched one-way ANOVA and Dunnett multiple-comparison correction: HBB TALEN R1/L1 *** p = 0.0007, R1/L2 ** p = 0.0016. (d) Representative immunoblot of human HBB, murine α-globin (Hba), and actin as calibrator for equal loading on day 6 of induced differentiation. Different same-blot, same-membrane hybridizations are separated by a hashed line. (e) Splice correction shown at the protein level as the ratios of HBB/Hba (±SD) on day 6 of induced differentiation, measured by immunoblots as shown in (d). Significant group-wise comparisons of ratios to no-electroporation controls for samples analyzed in triplicate by matched one-way ANOVA, assumed sphericity and Dunnett multiple-comparison correction: HBB TALEN R1/L1 * p = 0.0118, HBB TALEN R1/L2 ** p = 0.0054.