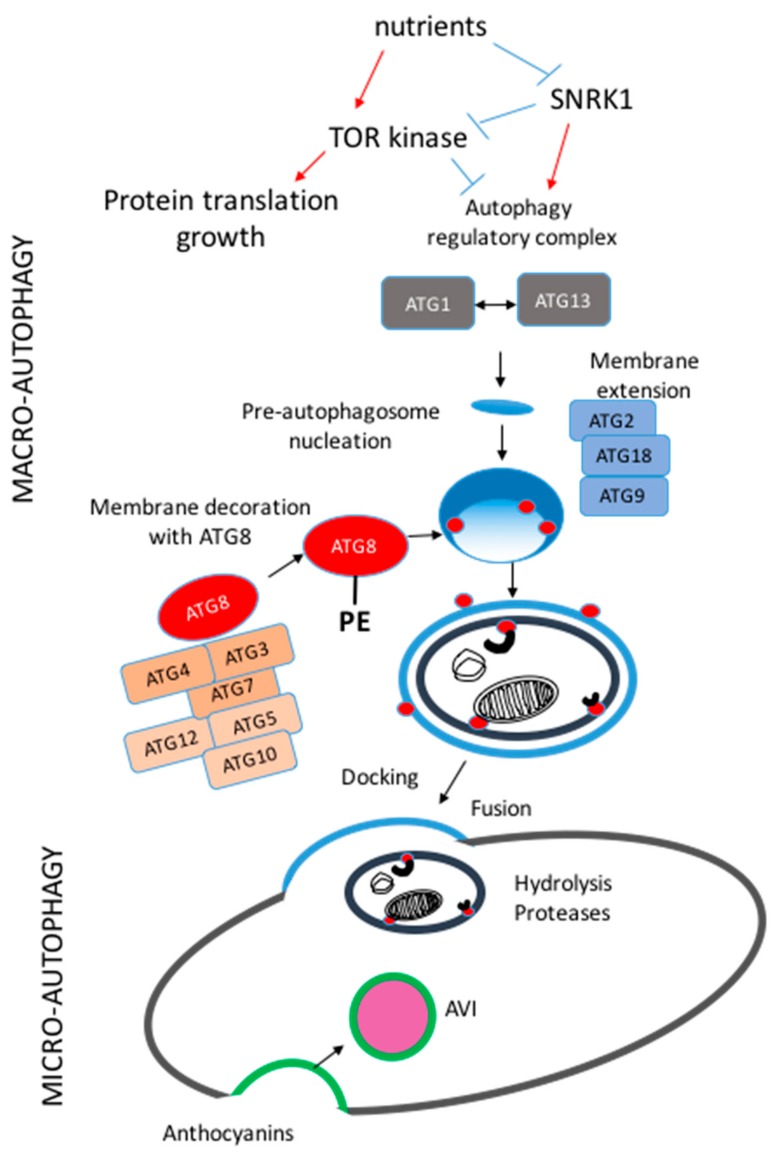
Figure 1.
Schematic representation of macro-autophagy and micro-autophagy pathways in plants. Nutrient availability controls the TOR (target of rapamycin) kinase activity that, in turn, regulates post-transcriptionally maro-autophagy through the phosphorylation of ATG1 and ATG13 (autophagy proteins 1 and 13). After nucleation of the pre-autophagosome structures, the autophagy ATG9, ATG18, and ATG2 proteins (in blue) are involved in the expansion of the membrane of the autophagosome. Several autophagy (ATG) proteins (in orange) involved in the conjugation of ATG8 to phosphatidyl-ethanolamine facilitate ATG8 anchorage to the membrane of the pre-autophagosome and per se autophagosome formation and enclosure. The ATG8-interacting motifs facilitate the capture of cargoes to be driven to the central vacuole for degradation. Micro-autophagy consists of the invagination of the tonoplast and participates in the formation of anthocyanin vacuole inclusions (AVI).