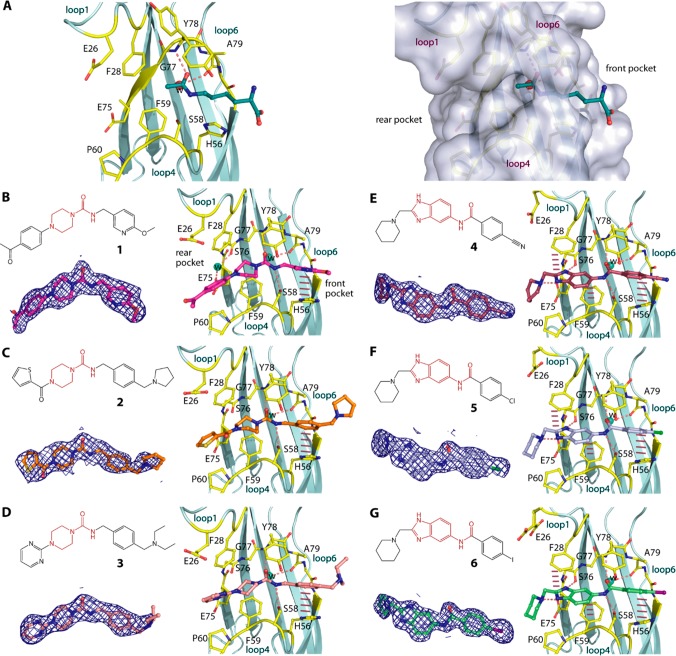
Figure 1.
Piperazine-urea and benzimidazole-amide derivatives and their binding modes in MLLT1. (A) The binding mode of acetyllysine in MLLT1 (pdb id: 6hpz) revealing potential front and rear pockets that could be targeted by small molecules. Chemical structures and the binding modes in MLLT1-complexed crystal structures of three piperazine-1-carboxamide 1, 2, and 3 (B–D; pdb ids: 6t1i, 6t1j, and 6t1l) and three benzimidazole-amide 4, 5, and 6 (E–G; pdb ids: 6t1m, 6t1n, and 6t1o). Dashed lines indicate hydrogen bonds, while bar, dashed lines are for potential aromatic π-stacking. Water molecules are shown in spheres. Electron density maps contoured at 1σ for the bound ligands are shown in the insets below the chemical structures in each panel. See Supplementary Figure S1 for omitted electron density maps.