Figure 7.
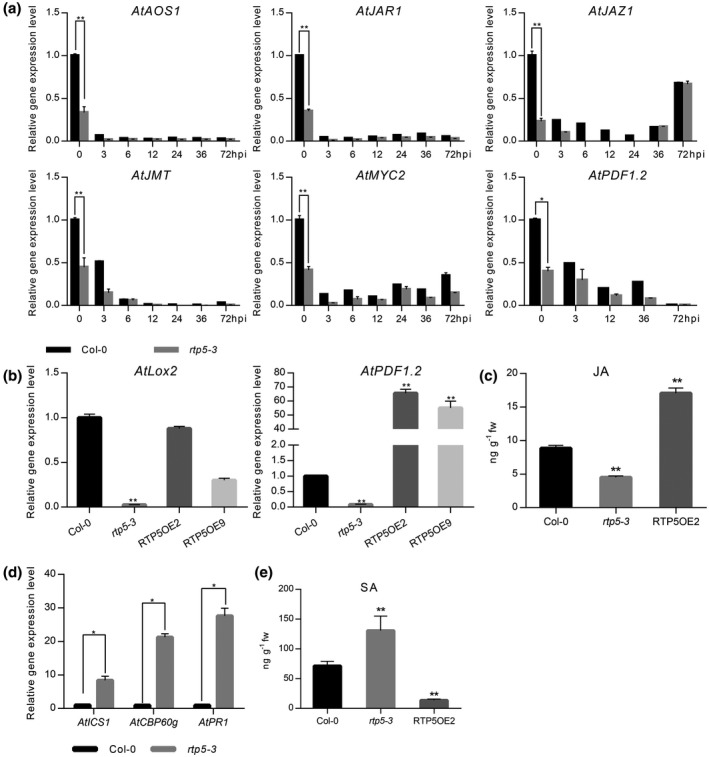
Deficiency of AtRTP5 suppressed jasmonic acid (JA) biosynthesis while activating the salicylic acid (SA) signalling pathway. (a) All examined JA signalling pathway marker genes were down‐regulated in rtp5‐3 compared with Arabidopsis thaliana Col‐0 during Phytophthora parasitica infection. Total RNA was obtained from detached leaves infected by P. parasitica at different times. (b) JA marker genes AtLox2 and AtPDF1.2 were down‐regulated in rtp5‐3 without infection, which were complemented by the overexpression of AtRTP5 (RTP5OE2 and RTP5OE9). (c) The endogenous JA contents of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 were determined by HPLC‐MS/MS with four biological replicates. The biosynthesis of JA was suppressed in rtp5‐3, but activated in RTP5OE2. (d) qRT‐PCR results showed that SA signalling pathway marker genes were up‐regulated in rtp5‐3. Total RNA was obtained from uninfected detached leaves of Col‐0 and rtp5‐3, respectively. (e) The endogenous SA contents of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 were determined by HPLC‐MS/MS with four biological replicates. SA was accumulated in rtp5‐3 and reduced in RTP5OE2, compared to Col‐0. The relative expression levels of tested genes in (a), (b) and (d) were normalized to AtUBC9. Three independent experiments were performed for each test of the gene expression levels. Statistical analysis was determined by Student’s t‐test between samples from two genotypes and determined by one‐way ANOVA followed by Tukey’s multiple comparison tests for samples from multiple genotypes.
