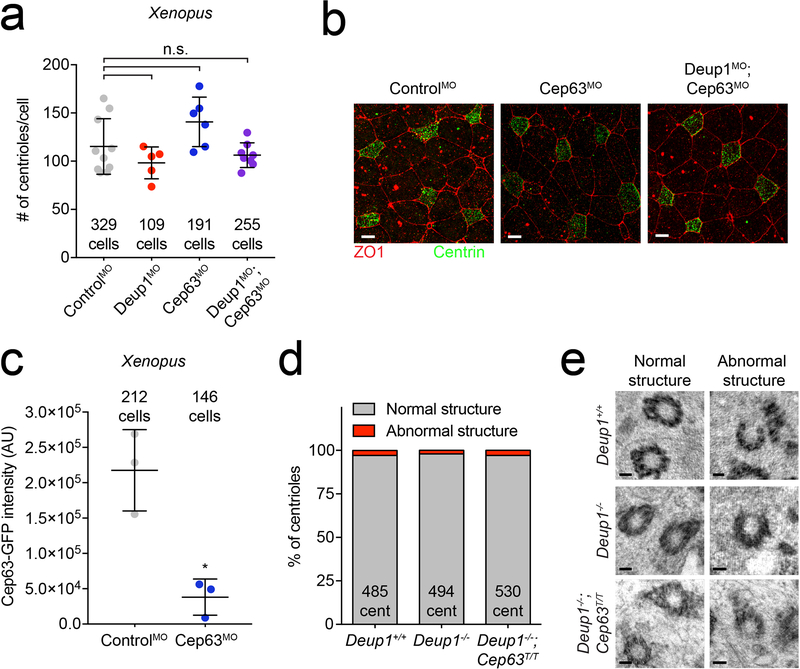
Extended Data Fig. 6: Deup1 and Cep63 are dispensable for centriole amplification in Xenopus.
(a) Quantification of centriole number in Xenopus epithelial cells treated with Cep63 or Deup1 and Cep63 morpholinos. Data from control and Deup1 morpholinos are from Extended Data 2d and shown for comparison. Points represent the average number of centrioles per cell in one embryo. n ≥ 5 embryos/genotype. The total number of cells analyzed per condition is indicated. P values, unpaired, two-tailed, Welch’s t-test. n.s. = not statistically significant (p > 0.05). Bars represent mean + SD.
(b) Representative immunofluorescence images from control, Cep63 or Deup1 and Cep63 morpholino treated Xenopus epithelial cells stained with tight junction marker, ZO1, and centriole marker, Centrin. Scale bars represent 10 μm.
(c) Quantification of Cep63-GFP intensity in control and Cep63 morpholino-treated Xenopus epithelial cells. Note, the Cep63 MO efficiently silenced expression of an mRNA encoding a morpholino-targetable Cep63 fused to GFP in Xenopus MCCs. n = 3 embryos/genotype. The total number of cells analyzed per condition is indicated. P values, unpaired, two-tailed, Welch’s t-test. * = p ≤ 0.05. Bars represent mean + SD.
(d) Quantification of the percent of centrioles with normal or abnormal structure in Deup1+/+, Deup1−/− and Deup1−/−; Cep63T/T ependymal cells. n > 30 cells/genotype. The total number of centrioles analyzed per genotype is indicated.
(e) Representative TEM images of normal and abnormal centrioles in Deup1+/+, Deup1−/− and Deup1−/−; Cep63T/T ependymal cells. Scale bar represents 100 nm.