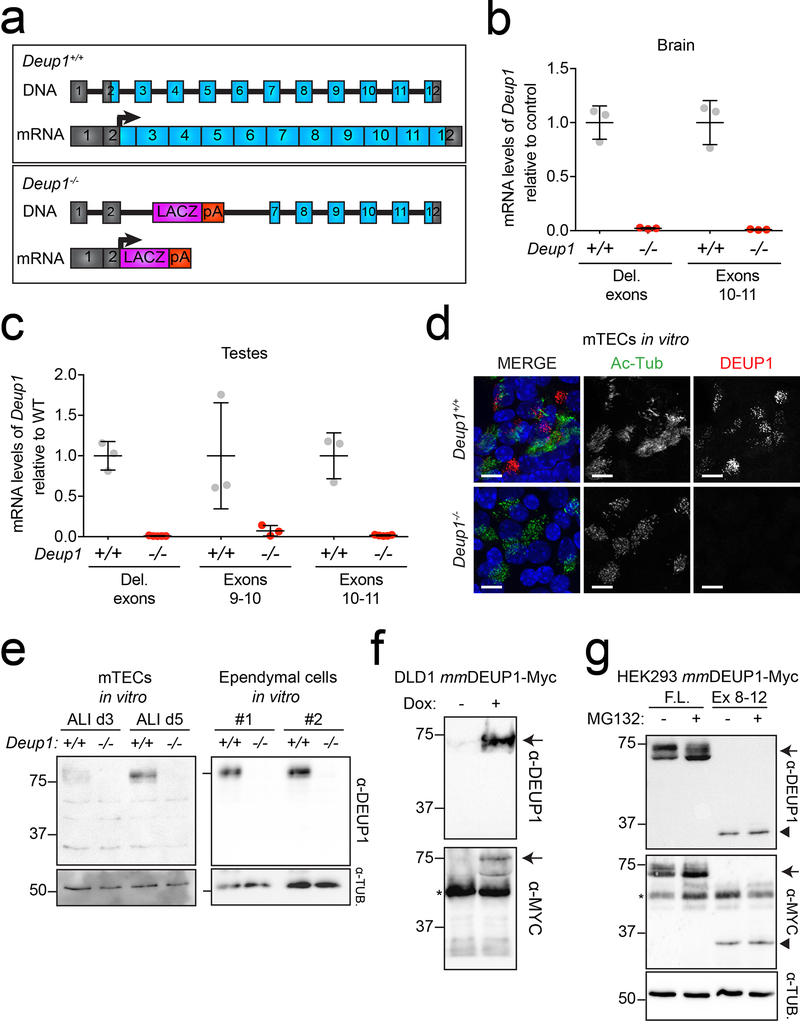
Extended Data Fig. 1: Deup1−/− mice lack Deup1 mRNA and detectable protein.
(a) A Deup1 knock-out was generated by replacing a region from within exon 2 to within exon 7 of the Deup1 gene with a LacZ reporter followed by a polyA sequence. (top) Schematic representation of the Deup1 gene and (bottom) mRNA structure in Deup1+/+ and Deup1−/− mice.
(b) RT-qPCR analysis of Deup1 mRNA levels in postnatal day 5 brain tissue. Deleted (Del.) exons denote primers designed to amplify exons 2–3 that are deleted in Deup1−/− mice. The average of Deup1+/+ samples was normalized to 1. n = 3 mice/genotype. Bars represent mean +/− SD.
(c) RT-qPCR analysis of testes in 2–5-month-old mice using 3 different primer sets. The average of Deup1+/+ samples was normalized to 1. n ≥ 3 mice/genotype. Bars represent mean +/− SD.
(d) Immunofluorescence images of mTECs at air-liquid interface (ALI) day 4. Scale bars represent 10 μm.
(e) Western blot of lysates from mTECs at ALI day 3 and 5, and two different ependymal cell cultures differentiated for 8 days. Membranes were probed with antibodies against DEUP1 and α-tubulin was used as a loading control. Full blot shown in Source Data.
(f) Immunoblot of DLD1 cells expressing doxycycline (dox) inducible mmDEUP1-Myc transgene. Membranes were probed with antibodies against DEUP1 or Myc. Arrow denotes the mmDEUP1-Myc protein and the asterisk shows endogenous Myc. Full blot shown in Source Data.
(g) Immunoblot of HEK293 cells expressing either full-length (F.L.) or exons 8–12 (Ex 8–12) of mmDEUP1 in the presence or absence of the proteasome inhibitor, MG132. MG132 was used to enable the detection of unstable protein fragments. Membranes were probed with antibodies against DEUP1 or Myc. α-tubulin was used as a loading control. Arrow denotes the full-length mmDEUP1-Myc protein, arrowhead denotes the exon 8–12 mmDEUP1-Myc protein and the asterisk shows endogenous Myc. Full blot shown in Sourc Data.