Abstract
An effective method to improve the blood supply of brain tissue is by angiogenesis, which is crucial for the prognosis of patients with cerebral ischemic stroke (CIS). Therefore, angiogenesis has been a focus of CIS research in recent years. The present study aimed to investigate the expression of microRNA (miR)-221 in patients with CIS and to explore the effect of miR-221 on endothelial cell function. The level of miR-221 was detected using reverse transcription-quantitative PCR (RT-qPCR). The relationship between miR-221 and phosphatase and tensin homolog (PTEN) was predicted and confirmed by bioinformatics and dual luciferase reporter assay. Cell viability, migration and invasion, and cell apoptosis were determined using MTT assay, Transwell assay and flow cytometry respectively. Tube formation in human umbilical vein endothelial cells (HUVECs) was determined by performing the tube formation assay. In addition, protein levels were measured using western blot analysis. The results of the current study indicated that miR-221 levels were significantly decreased in the peripheral blood of patients with CIS. PTEN was confirmed to be a direct target of miR-221. Downregulation of miR-221 significantly inhibited the function of HUVECs as evidenced by the decreased cell viability, migration and invasion with increased cell apoptosis and tube formation inhibition. miR-221 upregulation produced the reverse effects, whilst all the effects of miR-221 upregulation on HUVECs were reversed by PTEN overexpression. The PI3K/AKT pathway was identified to be involved in the regulation of miR-221 on HUVECs. In conclusion, miR-221 was downregulated in CIS patients, and it promoted the function of HUVECs by regulating the PTEN/PI3K/AKT pathway in vitro, suggesting the ability to promote angiogenesis. Therefore, miR-221 may be a novel and promising therapeutic target for CIS treatment.
Keywords: cerebral infarction, angiogenesis, human umbilical vein endothelial cells, microRNA-221, phosphatase and tensin homolog
Introduction
Cerebral ischemic stroke (CIS), a cerebrovascular disease with high incidence, disability rate and recurrence rate, seriously endangers human life and health accounting for 60–80% of all strokes (1,2). CIS imposes a burden on the patient's family and society (3). Therefore, in order to promote the recovery of nerve function, reduce the disability rate and improve the prognosis of CIS patients it is of great significance to explore new treatment methods for CIS.
Numerous studies have demonstrated that new blood vessels improve the blood supply of local brain tissue thereby providing nutritional support for neurons, saving sudden death neurons and improving the prognosis of CIS patients (4–6). Therefore, timely and effective promotion of neovascularization in the cerebral ischemic area has an important role in the recovery of neurological function of ischemic brain tissue. As an effective method to improve the blood supply of brain tissue, angiogenesis is crucial for the prognosis of patients with CIS (7–9) therefore has been the focus of CIS research in recent years. Angiogenesis refers to the proliferation, migration and remodeling of endothelial cells from original blood vessels as well as the formation of new blood vessels by budding to meet the biological function of local tissues (10). Angiogenesis serves an important role in various physiological and pathological processes such as wound healing, tissue regeneration, tumorigenesis and ischemia (11,12). Angiogenesis is a critical process of recovery from cerebrovascular disease (8,9).
MicroRNAs (miRNAs), a group of small non-coding RNAs, can regulate gene expression through binding to the 3′-untranslated regions (3′-UTRs) of the target mRNAs, and have key roles in the regulation of cell proliferation, differentiation and apoptosis (13,14). Currently, miRNAs have been identified as crucial regulatory factors that may serve as diagnostic biomarkers and/or therapeutic targets for human diseases. A large body of evidence indicates that miRNAs have important regulatory roles in angiogenesis (15–17). Increasing amounts of studies have demonstrated that miRNAs can participate in the development of CIS by regulating angiogenesis (18–20). miR-221 has been well studied in a variety of cancers (21–24) and has also been identified to be downregulated in CIS patients (25,26). However, to the best of our knowledge, the role and mechanism of miR-221 in CIS remains unclear.
In the present study, the expression pattern of miR-221 in the plasma of patients with CIS was investigated and in vitro experiments explored the effects and mechanisms of miR-221 on the function of human umbilical vein endothelial cells (HUVECs). Findings will hopefully provide a potential new target for the treatment of CIS.
Materials and methods
Clinical samples
A total of 20 samples of peripheral blood from 20 patients with CIS (13 males and 7 females; age range, 35 to 67 years) and 20 samples of peripheral blood from 20 healthy volunteers without any cerebrovascular diseases (12 males and 8 females; age range, 33 to 71 years) were collected at The Affiliated Hospital of Guizhou Medical University (Guizhou, China) from May 2016 to June 2018. Blood samples were centrifuged at 1,000 × g for 10 min at 4°C to obtain serum. The diagnosis of CIS was confirmed by computed tomography scan (CT) or magnetic resonance imaging scan (MRI) examinations. Inclusion criteria were as follows: i) Presentation of subjects within 72 h of the event; ii) National Institutes of Health Stroke Scale (NIHSS) score between 4 and 15 (27); and iii) APACHE II score evaluation <22, Cincinnati Score positive (28) for neurological symptoms at admission (including dysarthria and hemiparesis) and neuroimaging positive (CT or MRI positive). Exclusion criteria were patients with severe renal, liver or thyroid failure, acute infectious disease, rheumatic immune or hematologic disease, cancer or they had been taking lipid-lowering drugs within the last half of the year. The present study was approved by The Ethical Committee of the Affiliated Hospital of Guizhou Medical University and written informed consent was obtained from each patient.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were purchased from American Type Culture Collection. HUVECs were grown in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS (Hyclone; GE Healthcare Life Sciences) and the cells were incubated at 37°C and 5% CO2.
Cell transfection
HUVECs were transfected with 100 nM inhibitor control (5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma Co., Ltd.), 100 nM miR-221 inhibitor (5′-GAAACCCAGCAGACAAUGUAGCU-3′; Shanghai GenePharma Co., Ltd.), 50 nM mimic control (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; Shanghai GenePharma Co., Ltd.), 50 nM miR-221 mimic (sense, 5′-AGCUACAUUGUCUGCUGGGUUUC-3′ and antisense, 5′-AACCCAGCAGACAAUGUAGCUUU-3′; Shanghai GenePharma Co., Ltd.), 1 µg control-plasmid (cat. no. sc-437275; Santa Cruz Biotechnology, Inc.), 1 µg phosphatase and tensin homolog (PTEN)-plasmid (cat no. sc-400103-ACT; Santa Cruz Biotechnology, Inc.), 50 nM miR-221 mimic + 1 µg control-plasmid or 50 nM miR-221 mimic + 1 µg PTEN-plasmid for 48 h using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Transfection efficiency was detected by reverse transcription-quantitative PCR (RT-qPCR) 48-h following transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
To collect the total RNA from serum and cells, TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used according to the manufacturer's instructions. The PrimeScript™ RT reagent kit (Takara Bio, Inc.) was used to synthesize cDNAs following the manufacturer's instructions. The temperature protocol for the reverse transcription reaction was as follows: 50°C for 5 min and 80°C for 2 min. For qPCR, SYBR® Premix Ex Taq (Takara Bio, Inc.) was performed according to the manufacturer's protocol. The primer sequences used were as follows: U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3′; miR-221 forward, 5′-TGCGGAGCTACATTGTCTGCTGG-3′; and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′ and PTEN forward, 5′-GTCACTGCTTGTTGTTTGC-3′ and reverse, 5′-TTCTTTGTTGATAGCCTCCAC-3′. Thermocycling conditions were as follows: 10 min at 95°C followed by 37 cycles of 15 sec at 95°C and 40 sec at 55°C. Relative gene expression was quantified by the 2-∆∆Ct method (29) and normalized to U6 or GAPDH. Each experiment was performed in triplicate.
Western blot analysis
Total proteins were extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. To quantify the protein samples, a bicinchoninic acid protein quantitative kit (Thermo Fisher Scientific, Inc.) was used, according to the manufacturer's instructions. Then an equal amount of proteins (35 µg/lane) were separated by SDS-PAGE on 10% gels and transferred to polyvinylidene difluoride membranes. Subsequently, the membranes were blocked with 5% non-fat milk at room temperature for 1 h and then incubated with primary antibodies against PTEN (cat. no. 9188; 1:1,000; Cell Signaling Technology, Inc.), Bcl-2 (cat. no. 3498; 1:1,000; Cell Signaling Technology, Inc.), Bax (cat. no. 5023; 1:1,000; Cell Signaling Technology, Inc.), phosphorylated (p)-AKT (cat. no. 4060; 1:1,000; Cell Signaling Technology, Inc.), AKT (cat. no. 4685; 1:1,000; Cell Signaling Technology, Inc.) and β-actin (cat. no. 4970; 1:1,000; Cell Signaling Technology, Inc.) at 4°C overnight. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibody anti-rabbit IgG (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 2 h. Protein bands were visualized using an enhanced chemiluminescence kit (EMD Millipore) and the proteins were quantified by densitometry (Quantity One 4.5.0 software; Bio-Rad Laboratories, Inc.) with β-actin as the loading control.
MTT assay
Following specific treatments, cell viability was detected by MTT assay. The day before cell transfection, HUVECs were seeded into a 96-well plate (5×103 cells per well) and incubated at 37°C. Following transfection with or without inhibitor control, miR-221 inhibitor, mimic control, miR-221 mimic, miR-221 mimic + control-plasmid, or miR-221 mimic + PTEN-plasmid for 48-h, 20 µl MTT reagent was added to each well. Then the cells were incubated at 37°C for a further 4 h following which 150 µl dimethyl sulfoxide (DMSO) was applied in each well to dissolve the purple formazan crystals. Finally, the absorbance was detected at 490 nm wavelength using the FLUOstar® Omega Microplate Reader (BMG Labtech GmbH).
Flow cytometry
Following specific treatments, the apoptosis of HUVECs was analyzed using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit [cat. no.70-AP101-100; Hangzhou MultiSciences (Lianke) Biotech Co., Ltd.] according to the manufacturer's protocol. Briefly, 48 h following cell transfection, HUVECs were collected, washed with PBS, fixed in 70% ethanol at 4°C overnight and then stained with 5 µl Annexin V-FITC and 5 µl PI for 30 min in the dark at room temperature. A flow cytometer (BD FACS Aria; BD Biosciences) was used to analyze cell apoptosis and FlowJo software (version 7.6.1; FlowJo LLC) was used for data analysis. Experiments were repeated at least 3 times.
Transwell assay
In the present study, in vitro invasion and migration assays were performed using Transwell plates (pore size, 8 µm; BD Biosciences) with (invasion assay) or without Matrigel® (migration assay; BD Biosciences). The Matrigel pre-coating procedures were carried out at 37°C for 5 h. HUVECs (1×104 cells) diluted in serum-free DMEM were seeded into the upper chamber of the Transwell plates. Then 600 µl DMEM medium containing 20% FBS as the chemoattractant was added to the lower chamber. Following 48-h incubation, cells on the upper surface of the membrane were removed and the invasive or migratory cells on the lower surface of the membrane were fixed with methanol at room temperature for 30 min and then stained with 0.5% crystal violet at room temperature for 20 min. The membrane was counted in five randomly selected visual fields under an inverted light microscope (magnification, ×100; Olympus Corporation) using which the mean values were calculated.
Tube formation assay
Endothelial tube formation in HUVECs was determined by performing the tube formation assay. At 48-h following cell transfection, HUVECs (2×104 per well) were plated into the 96-well plates which were first pre-coated with 10 mg/ml Matrigel® at 37°C for 1 h (Becton, Dickinson and Company) and incubated in fully-supplemented DMEM for 24 h. To quantify tube formation, the tube length was calculated using ImageJ 1.38X software (National Institutes of Health).
Dual luciferase reporter assay
To predict the potential targets of miR-221, microRNA target site prediction software (www.microRNA.org; August 2010 Release) was used. The binding sites between the 3′-UTR of PTEN and miR-221 were identified. Then, a dual luciferase reporter assay was performed to confirm the binding sites between the 3′-UTR of PTEN and miR-221. The wild type (WT)-PTEN and mutant (MUT)-PTEN 3′-UTR of PTEN were cloned into a pmiR-RB-Report™ dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocol. To point-mutate the miR-221 binding domain on the 3′UTR of PTEN, a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) was performed in accordance with the manufacturer's protocol. HUVECs were co-transfected with 1 µg WT-PTEN or 1 µg MUT-PTEN and 50 nM miR-221 mimic or 50 nM mimic control using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in line with the manufacturer's protocols. Following 48 h incubation, a dual-luciferase assay system (Promega Corporation) was used to assess the luciferase activity. Luciferase activity was normalized to the Renilla luciferase activity.
Statistical analysis
Experiments were repeated for three times. SPSS 18.0 (SPSS, Inc.) was used to perform statistical analyses. Data are presented as the mean ± standard deviation. Comparison between two groups was made using Student's t-test or between multiple groups using one-way analysis of variance with Tukey's post hoc test. P<0.05 was considered to indicate statistical significance.
Results
miR-221 is significantly downregulated in the blood of patients with CIS
First, the level of miR-221 in the peripheral blood of patients with CIS and in healthy volunteers was detected using RT-qPCR. Results indicated that compared with the healthy volunteers, the level of miR-221 in the peripheral blood of patients with CIS significantly decreased (P<0.01; Fig. 1), indicating the important role of miR-221 in CIS.
Figure 1.

Expression of miR-221 in the blood of patient with CIS. The level of miR-221 in the peripheral blood of patients with CIS and in the healthy volunteers was detected using reverse transcription-quantitative PCR. **P<0.01 vs. healthy control. miR, microRNA; cerebral ischemic stroke, CIS.
PTEN is a direct target of miR-221
miRNA target site prediction software (microRNA.org) was used to identify the binding sites between miR-221 and the 3′-UTR of PTEN mRNA (Fig. 2A). The dual-luciferase reporter assay results indicated that compared with HUVECs co-transfected with mimic control and WT-PTEN, the luciferase activity of HUVECs co-transfected with miR-221 mimic and WT-PTEN was significantly reduced (P<0.01; Fig. 2B). However, compared with HUVECs co-transfected with mimic control and MUT-PTEN, there was no significant difference in the luciferase activity of HUVECs co-transfected with miR-221 mimic and MUT-PTEN (Fig. 2B). These results indicated that PTEN was a direct target of miR-221 in HUVECs.
Figure 2.
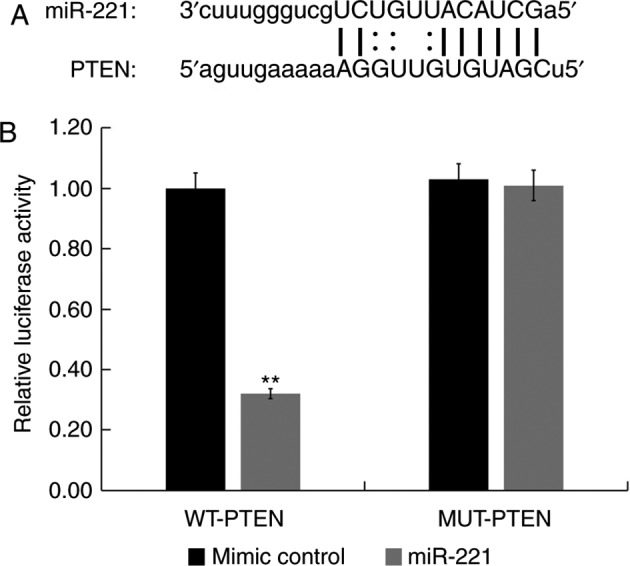
PTEN is a target of miR-221. (A) Binding sites between miR-221 and the 3′UTR of PTEN were predicted using miRNA target site prediction software. (B) Luciferase activity of a reporter containing a MUT-PTEN 3′UTR or a WT-PTEN were presented. All data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. respective mimic control. PTEN, phosphatase and tensin homolog; miR, microRNA; UTR, untranslated region; MUT, mutated; WT, wild-type.
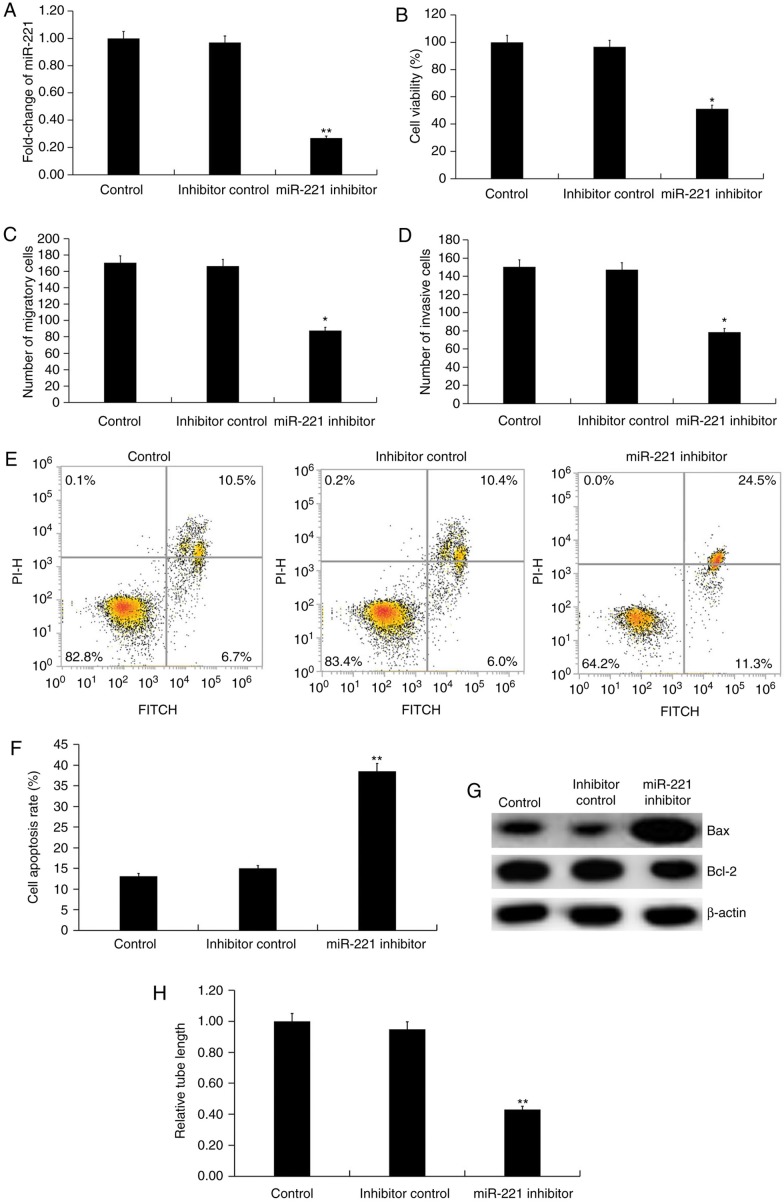
miR-221 downregulation inhibits the function of HUVECs
The effect of miR-221 downregulation on HUVECs was investigated using miR-221 inhibitor. Transfection with miR-221 inhibitor for 48 h significantly decreased the level of miR-221 in HUVECs (P<0.01; Fig. 3A). Further analysis indicated that compared with the inhibitor control, miR-221 inhibitor significantly decreased the cell viability (P<0.05; Fig. 3B), migration (P<0.05; Fig. 3C) and invasion ability (P<0.05; Fig. 3D) of HUVECs. Inhibition of miR-221 also induced cell apoptosis compared with the inhibitor control (P<0.01; Fig. 3E and F). The protein level of Bcl-2 was decreased by miR-221 inhibitor, while Bax protein expression was markedly enhanced in miR-221 inhibitor-transfected HUVECs compared with inhibitor control (Fig. 3G). To investigate the effect of miR-221 on the angiogenic abilities of endothelial cells, the tube formation of HUVECs was examined with findings demonstrating that miR-221 inhibitor repressed tube formation of HUVECs compared with the inhibitor control. (P<0.01; Fig. 3H).
Figure 3.
Effect of miR-221 inhibitor on HUVECs. (A) HUVECs were transfected with inhibitor control or miR-221 inhibitor for 48 h then, the level of miR-221 in HUVECs was detected using reverse transcription-quantitative PCR. (B) Cell viability was measured by an MTT assay. (C) Cell migration and (D) invasion were determined using Transwell and Matrigel assays, respectively. (E) Representative flow cytometry plots and (F) quantification of cell apoptosis was measured using flow cytometry. (G) Protein level of Bax and Bcl-2 was detected by western blot analysis. (H) Tube formation assay was used to determine cell tube formation ability. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. inhibitor control. miR, microRNA; HUVECs, human umbilical vein endothelial cells; PI, propidium iodide; FITC, fluorescein isothiocyanate.
miR-221 upregulation promotes the function of HUVECs
The effect of miR-221 upregulation on HUVECs was also investigated in the present study. HUVECs were transfected with control-plasmid, PTEN-plasmid, miR-221 mimic, miR-221 mimic + control-plasmid, or miR-221 mimic + PTEN-plasmid for 48 h. miR-221 mimic significantly increased the level of miR-221 in HUVECs compared with the mimic control group (P<0.01; Fig. 4A). PTEN-plasmid significantly enhanced the mRNA level of PTEN in HUVECs compared with the control-plasmid group (P<0.01; Fig. 4B). Compared with the mimic control group, miR-221 mimic significantly increased the cell viability (P<0.05; Fig. 4C), migration (P<0.05; Fig. 4D) and invasion ability (P<0.05; Fig. 4E) of HUVECs, and reduced cell apoptosis (P<0.01; Fig. 4F). In addition, miR-221 mimic markedly enhanced the protein level of Bcl-2 and inhibited Bax protein expression in HUVECs (Fig. 4G). In addition, miR-221 mimic significantly promoted the tube formation of HUVECs compared with the mimic control (P<0.01; Fig. 4H). More importantly, all the effects of miR-221 mimic on HUVECs were reversed by PTEN overexpression (Fig. 4).
Figure 4.
Effect of miR-221 mimic on HUVECs. (A) HUVECs were transfected with mimic control or miR-221 mimic for 48 h then the level of miR-221 was detected using RT-qPCR. (B) HUVECs were transfected with control-plasmid or PTEN-plasmid for 48 h, then the mRNA level of PTEN was detected using RT-qPCR. (C) HUVECs were transfected with mimic control, miR-221 mimic, miR-221 mimic + control-plasmid or miR-221 mimic + PTEN-plasmid for 48 h then cell viability was measured by an MTT assay. (D) Cell migration and (E) invasion were determined using Transwell and Matrigel assays, respectively. (F) Flow cytometry was used to analyze cell apoptosis. (G) Protein level of Bax and Bcl-2 was detected by western blot analysis. (H) Tube formation assay was used to determine cell tube formation ability. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. Mimic control; #P<0.05, ##P<0.01 vs. Mimic + control-plasmid. miR, microRNA; HUVECs, human umbilical vein endothelial cells; RT-qPCR, reverse transcription-quantitative PCR; PTEN, phosphatase and tensin homolog.
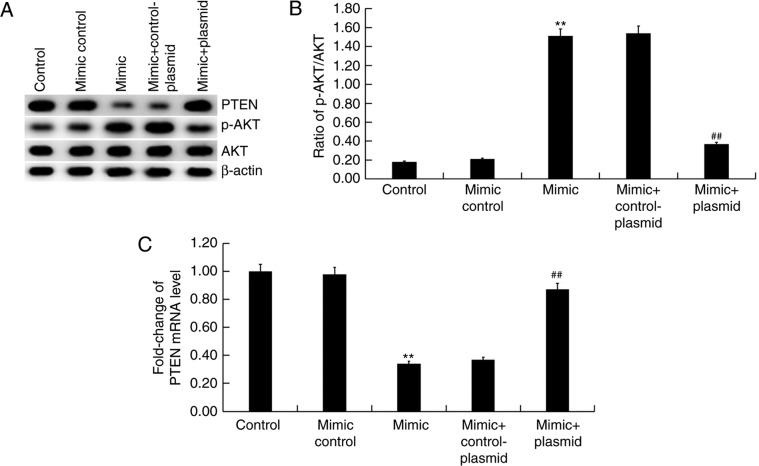
Effect of miR-221 on PTEN/PI3K/AKT pathway in HUVECs
Finally, the effect of miR-221 on PTEN/PI3K/AKT pathway was investigated. The results suggested that miR-221 mimic markedly inhibited PTEN protein expression and enhanced the level of AKT phosphorylation (Fig. 5A) compared with the mimic control group. Transfection with the miR-221 mimic significantly enhanced the ratio of p-AKT/AKT (Fig. 5B) compared with the mimic control group, in addition to significantly increasing the levels PTEN mRNA expression compared with the mimic control group (Fig. 5C). All the effects of miR-221 mimic on the expression of PTEN and p-AKT were reversed by PTEN overexpression (P<0.01; Fig. 5).
Figure 5.
Effect of miR-221 mimic on the PTEN/PI3K/AKT pathway in HUVECs. (A) Representative blots of HUVECs following transfection with mimic control, miR-221 mimic, miR-221 mimic + control-plasmid or miR-221 mimic + PTEN-plasmid. (B) Quantification of p-AKT protein level was measured by western blotting. (C) PTEN mRNA levels in HUVECs detected using reverse transcription-quantitative PCR. Data were presented as the mean ± standard deviation. **P<0.01 vs. Mimic control; ##P<0.01 vs. Mimic + control-plasmid. miR, microRNA; HUVECs, human umbilical vein endothelial cells; PTEN, phosphatase and tensin homolog; p, phosphorylated.
Discussion
The present study demonstrated that miR-221 was significantly downregulated in patients with CIS. Downregulation of miR-221 significantly inhibited the function of HUVECs as evidenced by the decreased cell viability, migration and invasion ability and tube formation with increased cell apoptosis. By contrast, miR-221 upregulation promoted cell viability, migration, invasion and tube formation of HUVECs and reduced cell apoptosis. In addition, miR-221 upregulation significantly inhibited PTEN expression and enhanced the phosphorylation of AKT in HUVECs. All the effects of miR-221 upregulation on HUVECs were eliminated by PTEN overexpression. The present findings indicated that miR-221 promoted the function of HUVECs and angiogenesis. Therefore, miR-221 might be a novel treatment target for CIS.
Currently, CIS seriously endangers human life and health (1,2). CIS recovery is a complex process, including inhibition of apoptosis, neuroinflammation, neurogenesis and angiogenesis (30,31). The angiogenic process of endothelial cells is critical for new blood vessel formation and blood flow recovery (9). In recent years, research on miRNA and angiogenesis in CIS has received increasing attention (16,32,33). miR-221, which belongs to the miR-221/222 cluster (34,35), has been well studied in a variety of cancers including hepatocellular carcinoma, prostate adenocarcinoma and colorectal carcinoma (21–22). miR-221 has also been identified to have an important role in tumor angiogenesis (24). Tsai et al (25) reported that miR-221 is significantly decreased in the serum of stroke patients and there is a 10.4-fold increase of stroke risk when miR-221 levels decrease. miR-221 has also been found to be downregulated in the cerebrospinal fluid of patients with CIS (26).
In the present study, consistent with the literature (25,26), it was identified that, the level of miR-221 in the peripheral blood of patients with CIS significantly decreased compared with the healthy volunteers, indicating that miR-221may serve a role during CIS. Consistent with previous studies (36–38), PTEN was confirmed to be a direct target of miR-221. PTEN is a dual lipid/protein phosphatase that has key roles in cell growth, migration and angiogenesis (39–41). However, contrary to the present results, Urbich et al (42) determined that miR-221 inhibits endothelial cell activity in vitro through indirectly regulating endothelial nitric oxide synthase expression (42). The reason for this discrepancy requires further research to elucidate the mechanisms. PTEN regulates the PI3K signaling pathway (40). A previous study revealed that human aortic smooth muscle cell-derived exosomes of miR-221 inhibit autophagy in HUVECs by modulating the PTEN/Akt signaling pathway (43). Therefore, to explore the mechanism by which miR-221 functioned on HUVECs, the effect of miR-221 on the PTEN/PI3K/AKT pathway was investigated in the present study. It was determined that miR-221 upregulation significantly inhibited the expression of PTEN in HUVECs, and as expected, the phosphorylation of AKT was significantly enhanced. All the effects of miR-221 mimic on HUVECs were reversed by PTEN overexpression.
This is only a preliminary study of the role of miR-221 in CIS therefore, further in-depth research should be performed to confirm its role. For example, the role of PTEN in angiogenesis should be investigated. The correlation between the expression levels of miR-221 and PTEN and the clinicopathological parameters of CIS patients needs to be explored. Finally, the function of miR-221 in CIS should be investigated in vivo. Future work will focus on these research areas.
In summary, the present study verified that miR-221 expression was downregulated in CIS patients (25,26). However, to the best of our knowledge, the present study was the first to demonstrate that miR-221 promoted the function of HUVECs by regulating the PTEN/PI3K/AKT pathway, thus promoting angiogenesis. Therefore, miR-221 might be a promising and novel therapeutic target for CIS treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science Foundation of China (grant no. H0928).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HP designed the current study, collected and analyzed the data, performed statistical analysis, searched the literature, and prepared the manuscript. HY and XX contributed to data collection and data interpretation. SHL contributed to statistical analyzes and interpreted the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethical Committee of the Affiliated Hospital of Guizhou Medical University, and written informed consent was obtained from by each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Middleton LE, Corbett D, Brooks D, Sage MD, Macintosh BJ, McIlroy WE, Black SE. Physical activity in the prevention of ischemic stroke and improvement of outcomes: A narrative review. Neurosci Biobehav Rev. 2013;37:133–137. doi: 10.1016/j.neubiorev.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Doyle KP, Simon RP, Stenzel-Poore MP. Neuropharmacology-special issue on cerebral ischemia mechanisms of ischemic brain damage-review article. Neuropharmacology. 2008;55:310–318. doi: 10.1016/j.neuropharm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dąbrowska-Bender M, Milewska M, Gołąbek A, Duda-Zalewska A, Staniszewska A. The impact of ischemic cerebral stroke on the quality of life of patients based on clinical, social and psychoemotional factors. J Stroke Cerebrovasc Dis. 2017;26:101–107. doi: 10.1016/j.jstrokecerebrovasdis.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 4.Zhu XF, Rao ML, Peng J, et al. Dynamis observed morphologic change of neuron and microcirculation in focal cerebral ischemia and reperfusion of rat. Chin J Clin Rehabil. 2002;6:1904–1905. (In Chinese) [Google Scholar]
- 5.Chen J, Choop M. Neurorestorative treatment of stroke: Cell and pharmacological approaches. NeuroRx. 2006;3:466–473. doi: 10.1016/j.nurx.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marti HJ, Bernaudin M, Bellail A, Schoch H, Euler M, Petit E, Risau W. Hypoxia-induced vascular endothelial growth factor expression precedes neovascularization after cerebral ischemia. Am J Patho1. 2000;156:965–976. doi: 10.1016/S0002-9440(10)64964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krupinski J, Kaluza J, Kumar P, Kumar S, Wang JM. Role of angiogenesis in patients with cerebral ischemic stroke. Stroke. 1994;25:1794–1798. doi: 10.1161/01.STR.25.9.1794. [DOI] [PubMed] [Google Scholar]
- 8.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 9.Ergul A, Alhusban A, Fagan SC. Angiogenesis: A harmonized target for recovery after stroke. Stroke. 2012;43:2270–2274. doi: 10.1161/STROKEAHA.111.642710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velazquez OC, Snyder R, Liu ZJ, Fairman RM, Herlyn M. Fibroblast-dependent differentiation of human microvaseular endothelial cells into capillary-like 3-dimensional networks. FASEB J. 2002;16:1316–1318. doi: 10.1096/fj.01-1011fje. [DOI] [PubMed] [Google Scholar]
- 11.Hillen F, Griffioen AW. Tumour vascularization: Sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26:489–502. doi: 10.1007/s10555-007-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy JA, Benjamin L, Zeng H, Dvorak AM, Dvorak HF. Vascular permeability, vasculax hyperpermeability and angiogenesis. Angiogenesis. 2008;11:109–119. doi: 10.1007/s10456-008-9099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Ranganathan K, Sivasankar V. MicroRNAs-Biology and clinical applications. J Oral Maxillofac Pathol. 2014;18:229–234. doi: 10.4103/0973-029X.140762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng N, Wang Z, Zhang Z, He X, Wang C, Zhang L. miR-487b promotes human umbilical vein endothelial cell proliferation, migration, invasion and tube formation through regulating THBS1. Neurosci Lett. 2015;591:1–7. doi: 10.1016/j.neulet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–15. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Wu F, Yang Z, Li G. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun. 2009;386:549–553. doi: 10.1016/j.bbrc.2009.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li LJ, Huang Q, Zhang N, Wang GB, Liu YH. miR-376b-5p regulates angiogenesis in cerebral ischemia. Mol Med Rep. 2014;10:527–535. doi: 10.3892/mmr.2014.2172. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhang Y, Zhang X, Zhang Y, Jiang Y, Xiao X, Tan J, Yuan W, Liu Y. MicroRNA-433 inhibits the proliferation and migration of HUVECs and neurons by targeting hypoxia-inducible factor 1 alpha. J Mol Neurosci. 2017;61:135–143. doi: 10.1007/s12031-016-0853-1. [DOI] [PubMed] [Google Scholar]
- 20.Liang Z, Chi YJ, Lin GQ, Luo SH, Jiang QY, Chen YK. MiRNA-26a promotes angiogenesis in a rat model of cerebral infarction via PI3K/AKT and MAPK/ERK pathway. Eur Rev Med Pharmacol Sci. 2018;22:3485–3492. doi: 10.26355/eurrev_201806_15175. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H, Xu N, Xie Y. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One. 2014;9:e109347. doi: 10.1371/journal.pone.0109347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q, Peskoe SB, Ribas J, Rafiqi F, Kudrolli T, Meeker AK, De Marzo AM, Platz EA, Lupold SE. Investigation of miR-21, miR-141 and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate. 2014;74:1655–1662. doi: 10.1002/pros.22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Wang W, Zhou C, Xi W, Yuan L, Chen X, Li Y, Yang A, Zhang J, Wang T. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 2015;36:3763–3773. doi: 10.1007/s13277-014-3017-3. [DOI] [PubMed] [Google Scholar]
- 25.Tsai PC, Liao YC, Wang YS, Lin HF, Lin RT, Juo SH. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J Vasc Res. 2013;50:346–354. doi: 10.1159/000351767. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res. 2014;5:711–718. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 27.Heldner MR, Zubler C, Mattle HP, Schroth G, Weck A, Mono ML, Gralla J, Jung S, El-Koussy M, Lüdi R, et al. National Institutes of Health stroke scale score and vessel occlusion in 2152 patients with acute ischemic stroke. Stroke. 2013;44:1153–1157. doi: 10.1161/STROKEAHA.111.000604. [DOI] [PubMed] [Google Scholar]
- 28.Powers WJ, Rabinstein AA, Ackerson T, Adeoye OM, Bambakidis NC, Becker K, Biller J, Brown M, Demaerschalk BM, Hoh B, et al. 2018 Guidelines for the early management of patients with acute ischemic stroke: A guideline for healthcare professionals from the American heart association/American stroke association. Stroke. 2018;49:e46–e110. doi: 10.1161/STR.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Attanasio S, Schaer G. Therapeutic angiogenesis for the management of refractory angina: Current concepts. Cardiovasc Ther. 2011;29:e1–e11. doi: 10.1111/j.1755-5922.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- 31.Chopp M, Zhang ZG, Jiang Q. Neurogenesis, angiogenesis, and MRI indices of functional recovery from stroke. Stroke. 2007;38(2 Suppl):S827–S831. doi: 10.1161/01.STR.0000250235.80253.e9. [DOI] [PubMed] [Google Scholar]
- 32.Yuan Y, Zhang Z, Wang Z, Liu J. MiRNA-27b regulates angiogenesis by targeting ampk in mouse ischemic stroke model. Neuroscience. 2019;398:12–22. doi: 10.1016/j.neuroscience.2018.11.041. [DOI] [PubMed] [Google Scholar]
- 33.Yin KJ, Hamblin M, Chen YE. Angiogenesis-regulating microRNAs and ischemic stroke. Curr Vasc Pharmacol. 2015;13:352–365. doi: 10.2174/15701611113119990016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ, Tang JH, Zhao JH. miR-221/222: Promising biomarkers for breast cancer. Tumour Biol. 2013;34:1361–1370. doi: 10.1007/s13277-013-0750-y. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu Z, Cao X, Chen P, Liu Z, et al. A microRNA 221-and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong ZH, Zhou F, Shi C, Xiang T, Zhou CK, Wang QQ, Jiang YS, Gao SF. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24:9. doi: 10.1186/s11658-018-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren Y, Yang M, Ma R, Gong Y, Zou Y, Wang T, Wu J. Microcystin-LR promotes migration via the cooperation between microRNA-221/PTEN and STAT3 signal pathway in colon cancer cell line DLD-1. Ecotoxicol Environ Saf. 2019;167:107–113. doi: 10.1016/j.ecoenv.2018.09.065. [DOI] [PubMed] [Google Scholar]
- 38.Cortés-Garcia JD, Briones-Espinoza MJ, Vega-Cárdenas M, Ruíz-Rodríguez VM, Mendez-Mancilla A, Gómez-Otero AE, Vargas Morales JM, García-Hernández MH, Portales-Pérez DP. The inflammatory state of adipose tissue is not affected by the anti-inflammatory response of the A2a-adenosine system and miR-221/PTEN. Int J Biochem Cell Biol. 2018;100:42–48. doi: 10.1016/j.biocel.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: The long and the short of it. Trends Biochem Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serra H, Chivite I, Angulo-Urarte A, Soler A, Sutherland JD, Arruabarrena-Aristorena A, Ragab A, Lim R, Malumbres M, Fruttiger M, et al. PTEN mediates Notch-dependent stalk cell arrest in angiogenesis. Nat Commun. 2015;6:7935. doi: 10.1038/ncomms8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Wang Z, Hu X, Wan T, Wu H, Jiang W, Hu R. Human aortic smooth muscle cell-derived exosomal miR-221/222 inhibits autophagy via a PTEN/Akt signaling pathway in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2016;479:343–350. doi: 10.1016/j.bbrc.2016.09.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.