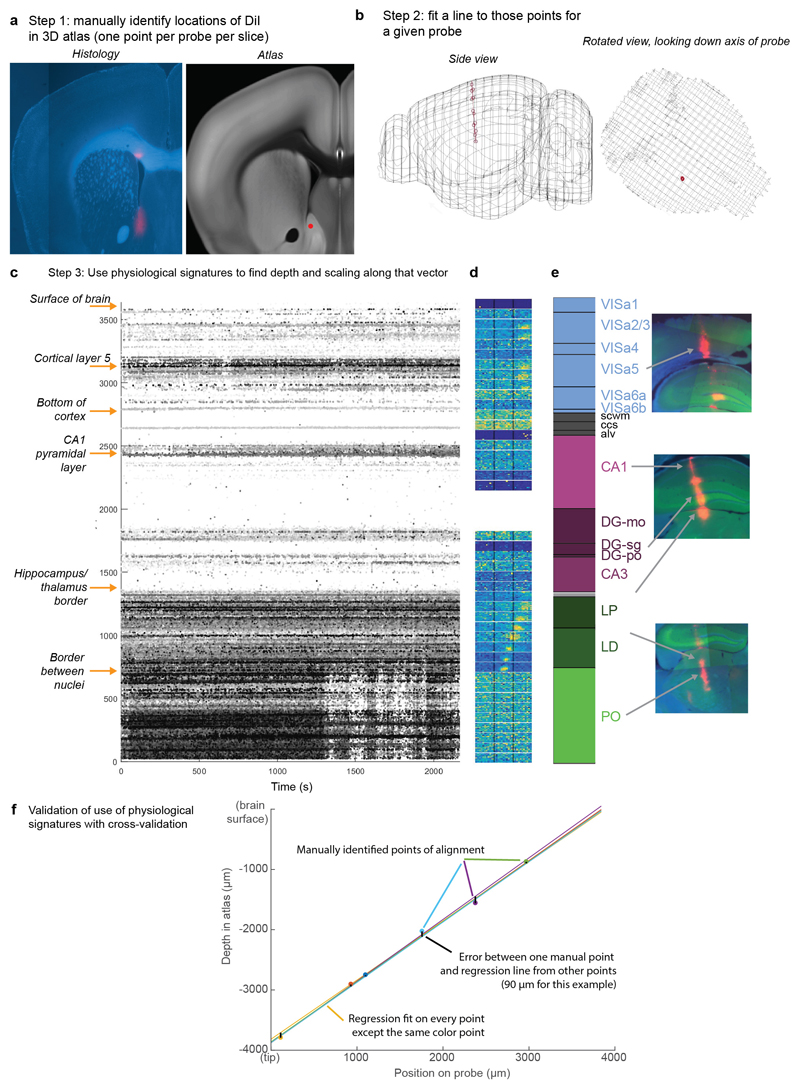
Extended Data Figure 2. Method for histological alignment.
a, Prior to insertion, probes are dipped in DiI. The brain is sliced and imaged, and locations of each probe’s DiI spots are manually identified on the Allen CCF atlas (15.1 ± 6.9 per probe). When multiple penetrations were performed in a single brain, their tracks are sufficiently far apart to avoid confusion. b, A vector is fit to the probe track using total least squares linear regression. The median distance of individual points from this vector is 39.3 μm, providing an estimate of lateral displacement error. c, To fit the longitudinal mapping from recording sites to brain locations, we used landmarks easily detectable by their electrophysiological signatures (arrows, left), linearly interpolating the location of sites between these landmarks. d, visual receptive fields served as a post-hoc check on correct alignment, but were not used to estimate track location. Each horizontally elongated plot with two vertical black lines indicates the responsiveness of all spikes recorded in an 80 μm depth bin to flashed white squares at varying locations on the three screens (see Methods, receptive field mapping). Colormap brightness is proportional to spike rate, independently scaled for each map. e, areas assigned for each recording site. Right: example DiI traces in slices corresponding to these locations. f, Example of cross-validation procedure to assess error in longitudinal alignment. For each point, the longitudinal mapping was recomputed excluding this point, and the distance from this point to the mapping fit to other points provides an estimate of longitudinal alignment error. Brain diagrams were derived from the Allen Mouse Brain Common Coordinate Framework (version 3 (2017); downloaded from http://download.alleninstitute.org/informatics-archive/current-release/mouse_ccf/).