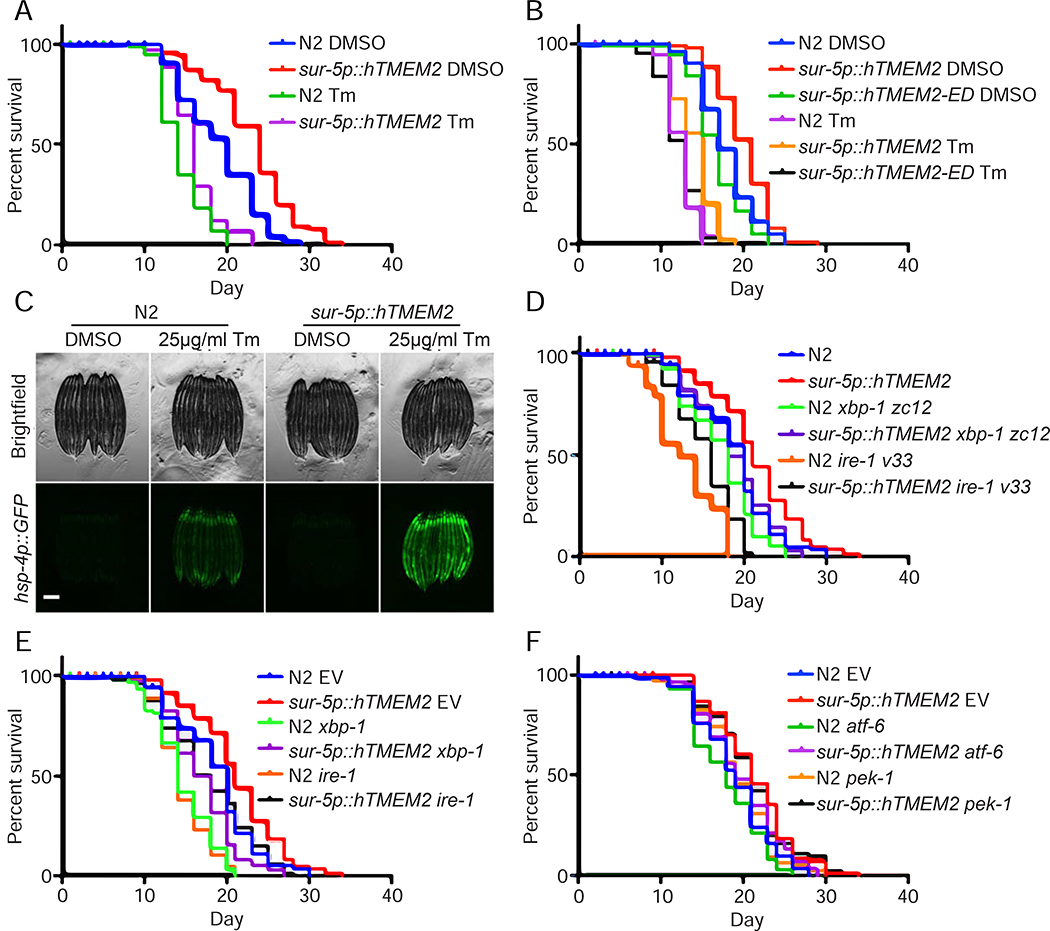
Figure 5:
hTMEM2 overexpression extends lifespan in C. elegans independent of canonical UPRER pathways. A) Lifespans were measured in Wildtype (N2) and sur-5p::hTMEM2 worms grown on EV RNAi on 1% DMSO and 25μg/ml Tunicamycin (Tm) from Day 1 (D1) as described in STAR METHODS. Data is representative of four independent trials. B) Lifespans were measured in Wildtype (N2), sur-5p::hTMEM2, and an enzymatic dead version of hTMEM2 (sur-5p::hTMEM2-ED, carrying R265C, D273N, D286N mutations; this overexpression line is an extrachromosomal array) using similar methods as A. Data is representative of three independent trials. C) Fluorescent micrographs of Wildtype (N2) and sur-5p::hTMEM2 animals expressing the UPRER reporter, hsp-4p::GFP. Animals were treated with DMSO or 25μg/ml Tunicamycin (Tm) at L4, and imaged at D1 as described in STAR METHODs. Data is representative of four independent trials. D) Lifespans were measured in Wildtype and sur-5p::hTMEM2 animals carrying either Wildtype alleles of xbp-1 and ire-1 or mutant alleles, xbp-1(zc12) or ire-1(v33), on EV RNAi. Data is representative of three independent trials. E) Lifespans were measured in Wildtype and sur-5p::hTMEM2 animals grown on EV, xbp-1, or ire-1 RNAi from hatch. Data is representative of three independent trials. F) Lifespans were measured in Wildtype and sur-5p::hTMEM2 animals grown on EV, atf-6, or pek-1 RNAi from hatch. Data is representative of two independent trials. All statistics for lifespans were performed using Log-Rank (Mantel-Cox) test using PRISM, and are available in Supplemental Table 2.