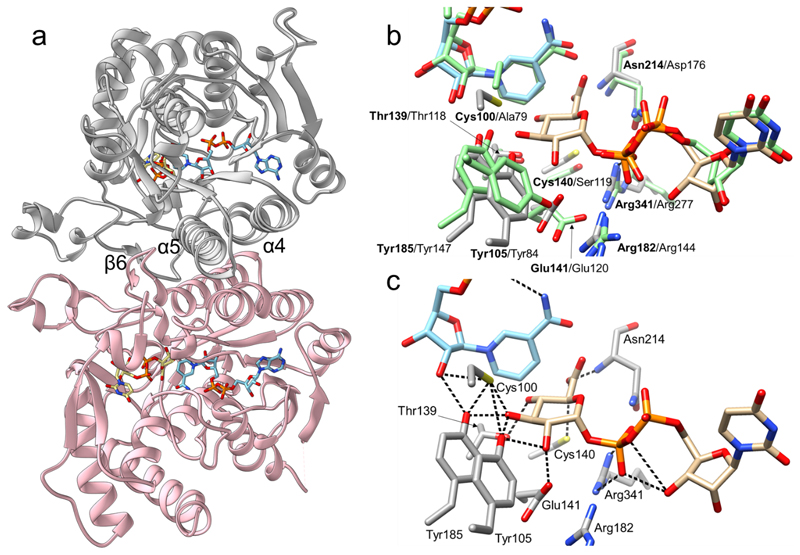
Fig. 2. Crystallographic analysis of the three-dimensional structure of UAXS from Arabidopsis thaliana.
(a) The UAXS subunit features the typical SDR fold. The enzyme is a functional dimer with chain A shown in grey and chain B shown in pink. The dimer interface of UAXS is composed of helices α4 (residues 109-129) and α5 (residues 181-202). There are additional dimer contacts from strand β6 (residues 170-179) and from an extended loop. NAD+ is represented in cyan carbons and UDP-GlcA (2) in beige carbons. Oxygen atoms are in red, nitrogen atoms in blue, phosphorus atoms in orange, and sulfur atoms in yellow. (b) Structural comparison/overlay of the active sites of UAXS and human UXS in complex with UDP and NAD+ (PDB entry 2B69; light green carbon atoms). Most of the residues are conserved and have similar conformations. The main changes are replacements of Cys100 and Cys140 of UAXS with Ala79 and Ser119 in human UXS (Supplementary Figure 2). (c) A model for the UAXS Michaelis complex (enzyme:NAD+:UDP-GlcA) was built by combining the ligands from two different structures; the wild-type enzyme bound to NAD+ and UDP (Supplementary Figure 7), and the C100A mutant bound to NADH and UDP-GlcA (Supplementary Figure 8). Enzyme residues are displayed in light grey carbon atoms, UDP-GlcA is represented with beige carbon atoms, NAD+ with cyan carbons. Cys100 was modelled in a rotameric conformation suitable to interact with 3’-OH of the sugar substrate. Possible H-bond interactions between the labelled residues and the ligands are shown as black dashed lines.