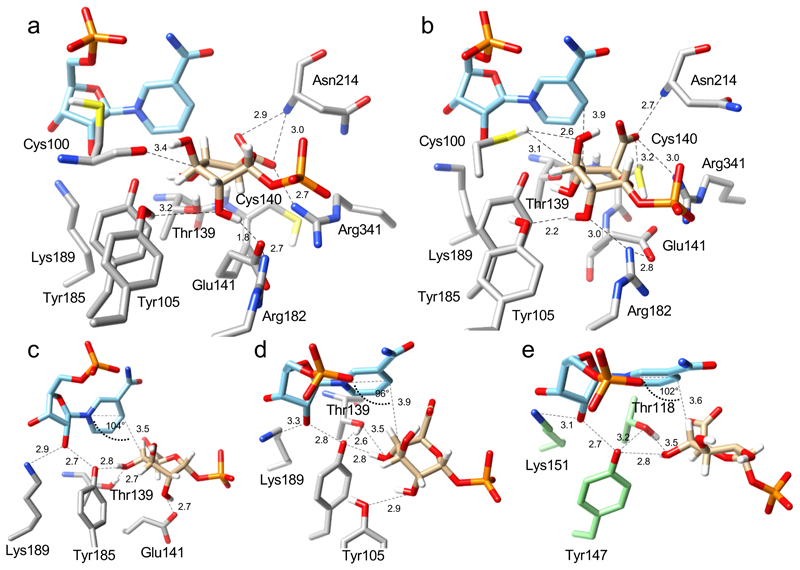
Fig. 3. Positioning of the UDP-GlcA substrate for catalytic oxidation and ring opening at the active site of UAXS, revealed by QM/MM calculations.
The QM/MM calculations were performed on selected, highly representative snapshots from the MD simulations. (a) Close-up structure and key interactions for the proposed Michaelis complex; the ternary NAD+:UDP-GlcA:enzyme complex is from Replica 1 (Supplementary Video 1). The pyranosyl ring of GlcA adopts a 1S5 conformation. (b) Close-up structure and key interactions for an alternative ternary complex, prominently present in MD simulations from Replica 2 (Supplementary Video 2). The pyranosyl ring of GlcA adopts a 1C4 conformation. (c), (d) Further close ups, taken from panels a and b, respectively, to highlight the catalytic interactions for oxidation of the C4’ alcohol group, via hydride transfer to NAD+ and proton transfer to Tyr185, and sugar ring opening, via deprotonation of the 2’-hydroxy group. Note: the orientations from panels a and b were adapted for better viewing. (e) Modeled Michaelis complex of human UXS with bound UDP-GlcA (2) and NAD+.25