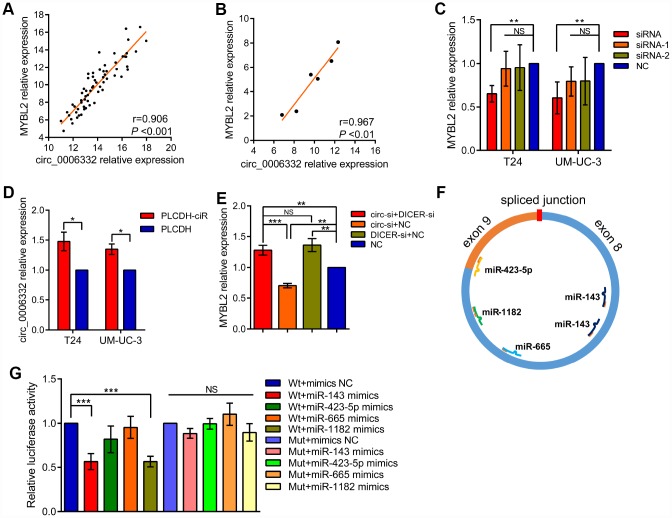
Figure 4.
Circ_0006332 increases MYBL2 expression by sponging miR-143. (A–B) Correlation analysis shows that circ_0006332 levels correlate with MYBL2 expression in bladder cancer tissues and cell lines including 5637, T24, J82, UM-UC-3, TSCCUP and SV-HUC-1. (C) QRT-PCR analysis shows MYBL2 mRNA levels in bladder cell lines transfected with siRNA, siRNA-1, siRNA-2 and negative control against circ_0006332. As shown, MYBL2 expression is significantly decreased in circ_0006332 knockdown bladder cancer cells, but is not downregulated in cells transfected with siRNA-1 and siRNA-2. (D) QRT-PCR analysis shows MYBL2 mRNA levels in control and circ_0006332 overexpressing bladder cancer cells. Bladder cancer cells were transfected with PLCDH-ciR, the cloning vector with circ_0006332 or control vector, PLCDH. As shown MYBL2 mRNA levels are higher in cells overexpressing circ_0006332 compared with controls. (E) QRT-PCR analysis shows MYBL2 mRNA levels in bladder cancer cells transfected with negative control and siRNA against Dicer. As shown, Dicer knockdown cells eliminate the effects of circ_0006332 on MYBL2 expression. DICER-si, siRNA of DICER; circ-si, siRNA of circ_0006332 (F) MiRwalk and CircInteractome predict 4 miRNAs most likely bind to circ_0006332. As shown, circ_0006332 potentially bind to miR-143, miR-423-5p, miR-665, miR-1182 at position of 180-186 and 222-227, 458-464, 343-349, 382-395 respectively. (G) Results of the dual luciferase reporter assays confirm that miR-143 binds to wild type circ_0006332 (Wt) but do not bind to mutant circ_0006332 (Mut). Note: All experiments were repeated thrice and data were represented as mean ± SD; NS, no significance; *P < 0.05, **P < 0.01, ***P < 0.001.