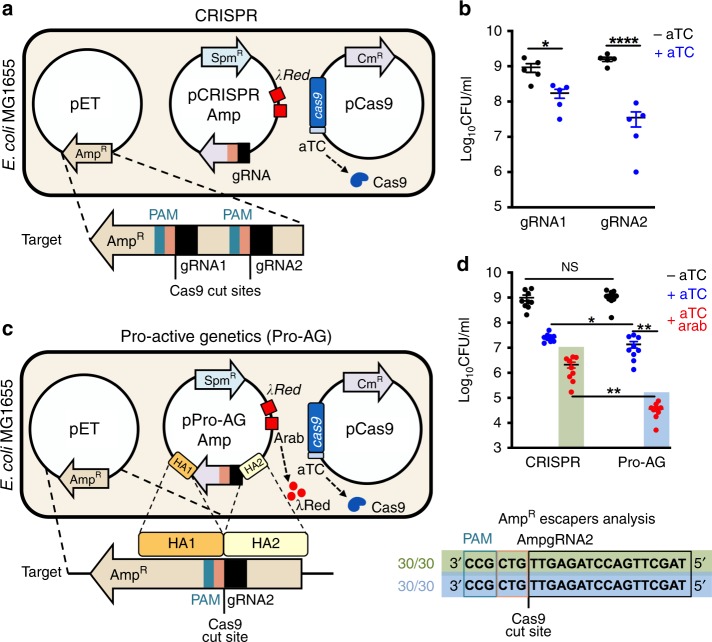
Fig. 1. Pro-active genetics (Pro-AG) is ~ 100× more efficient than CRISPR-control for targeting antibiotic resistance conferred by a high copy plasmid.
a and c Schematic of CRISPR-control- a and Pro-AG- c mediated editing of beta-lactamase gene (bla) encoded on a high copy plasmid (pET) conferring resistance to ampicillin (AmpR, tan arrow) in E. coli MG1655. gRNAs were initially expressed from a low copy plasmid (pCRISPR Amp) maintained under spectinomycin (Spm) selection and using the constitutive tet promoter to express the gRNA. A second low copy plasmid, pCas9, maintained under chloramphenicol (CmR) selection, encodes Cas9 under control of an anhydrotetracycline (aTc) inducible promoter. a CRISPR-control configuration: two gRNAs (gRNA1 and gRNA2 carried on the pCRISPR Amp plasmid) were tested for Cas9-mediated targeting at two different locations in the bla gene. The Cas9/gRNA1 and Cas9/gRNA2-induced cleavage sites, and the protospacer adjacent motif (PAM) are indicated as well as a gene cassette carrying the λRed enzymes. b Recovery of AmpR colony-forming units (Log10CFU/ml) following CRISPR-mediated targeting of the bla gene with gRNAs 1 or 2 in the absence (− aTc, black dots) or presence (+ aTc, blue dots) of aTc-induced Cas9 expression. c Pro-AG configuration: the gRNA2 expression cassette flanked by bla (AmpR) homology arms (HA1 and HA2) that directly abut the gRNA2 cleavage site was incorporated into pCRISPR Amp. The Cas9/gRNA2-induced cleavage site, the protospacer adjacent motif (PAM), and the HA1 (dark yellow box), and HA2 (light yellow box) homology arms are indicated. Also carried on plasmids pCRISPR Amp and pPro-AGAmp are the recombinogenic λRed enzymes (λRed), which can be induced with l-arabinose (arab). d Recovery of CFU on Amp plates following CRISPR-control versus Pro-AG-mediated targeting of the bla gene. In this and subsequent panels, abbreviations for Cas9 induction are as in panel b; induction of λRed enzymes: (+ aTc + arab: red dots); CRISPR (green shaded box) and Pro-AG (blue shaded box) treatments are highlighted. Plasmids sequenced from AmpR colonies (CRISPR, green box and Pro-AG, blue box) displayed unaltered gRNA target sites in all clones analyzed (30/30). Data in b and d are plotted as the mean ± SEM, representing three independent experiments performed in triplicate and analyzed by Student’s t test. N.S. = not significant (P > 0.05) *P < 0.05; **P < 0.01; ****P < 0.0001. Source Data are available in the Source Data file.