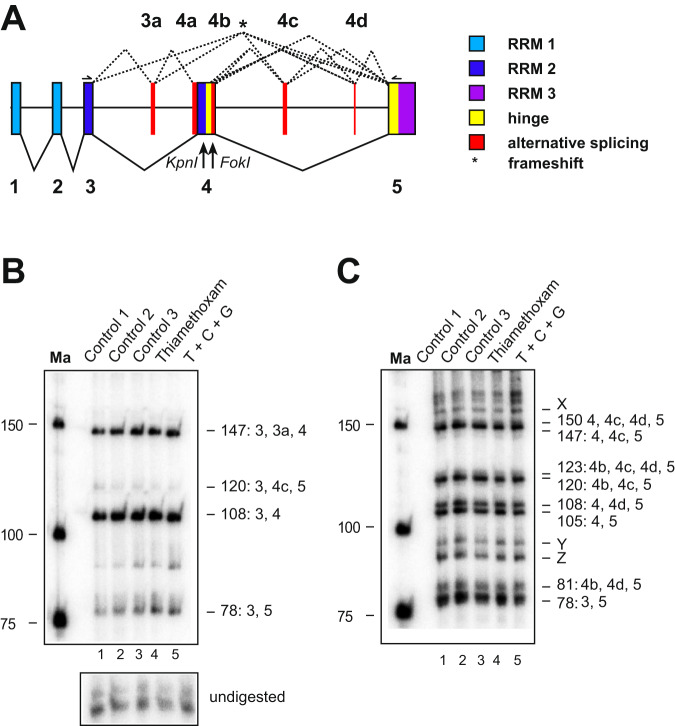
Figure 5.
Apis mellifera elav alternative splicing in brains of worker bees is unaffected by thiamethoxan, carbendazim and glyphosate. (A) Gene structure of Apis mellifera elav depicting color-coded functional protein domains with constant exons (1–5, bottom, solid lines) and alternative splicing exons (3a and 4a–d, top, dashed lines). RNA Recognition Motiv 1 (RRM1): light blue, RRM2: dark blue, RRM3: purple, hinge region: red and alternatively spliced parts in red. KpnI and FokI restriction sites used to separate isoforms are indicated below the gene model. An asterisk indicates isoforms that encode truncated proteins by introducing a frameshift. (B,C) Denaturing polyacrylamide gels (6%) showing the alternative splicing pattern of elav by digestion of a 5′ (B) or 3′ (C) 32P labeled RT-PCR product with KpnI (B) and FokI (C) in control bees dissected immediately after collection (Control 1), control bees fed with water and sucrose for 24 h (Control 2) and control bees injected with water (Control 3) compared to bees injected with thiamethoxam (1 µM) and bees injected with a mixture of thiamethoxam (1 µM, T), carbendazim (2 mM, C) and glyphosate (32 mM, G) 24 h prior dissection. Samples were run on 6% polyacrylamide gel. Ma: DNA marker. The undigested PCR product is shown at the bottom.