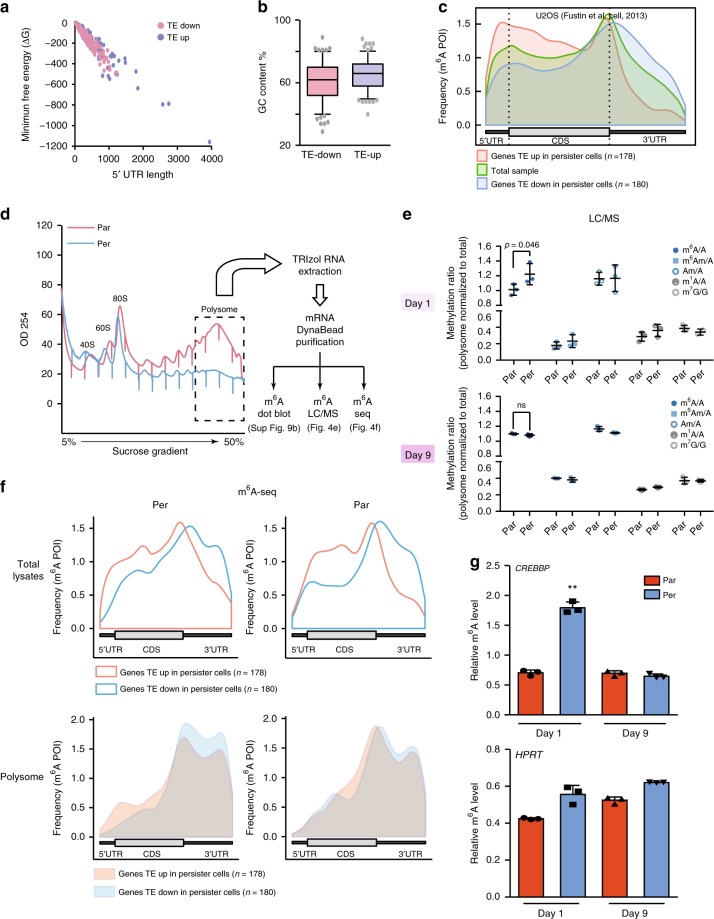
Fig. 4. N6-methyladenosine (m6A) is enriched in the mRNAs translationally upregulated in persister cells.
a 5′-UTR minimum free energy and length of the mRNAs translationally upregulated and top 180 downregulated in persister cells. b GC content of the mRNAs translationally upregulated and top 180 downregulated in persister cells. TE: translation efficiency. c The distribution of m6A peaks for mRNA upregulated (red) and downregulated (blue) at the translational level in persister cells. The whole population of mRNAs was plotted in green as a control. d Heavy polysome-bound mRNAs were extracted and purified for m6A dot plot assay, m6A LC/MS-MS assay and m6A RNA immunoprecipitation followed by RNA sequencing (m6A-seq) in persister vs. parental cells. e Liquid chromatography–mass spectrometry (LC/MS-MS) nucleoside modification analysis. Total RNAs were purified by poly(A) enrichment and were subjected to digestion. Triplicate samples were then subjected to LC/MS-MS quantification. Each type of RNA methylation was first normalized to non-methylated nucleotide base (A or G) and polysome RNA methylation was then normalized to total RNA methylation (input). m6A: 6-methyladenosine; m6Am: N6,2’-O-dimethyladenosine; Am: 2’-O-methyladenosine; m1A: 1-methyladenosine; m7G: 7-methylguanosine; A: adenosine; G: guanosine (n = 3, unpaired t-test, ns: nonsignificant). f Metagene profiles of enrichment of m6A modifications across mRNAs corresponding to translationally upregulated transcripts and the top 180 translationally downregulated transcripts in parental and persister cells. Top panel, metagene profiles of m6A modifications of total lysate from parental or persister cells; bottom panel: metagene profiles of m6A modifications of heavy polysome fractions from parental or persister cells. CDS, coding sequence. g RT-qPCR quantification of m6A enrichment in polysome-bound CREBBP mRNA and HPRT mRNA at indicated time points (n = 3, **p-value < 0.01, unpaired t-test). The raw data of e and g are available in Source Data.