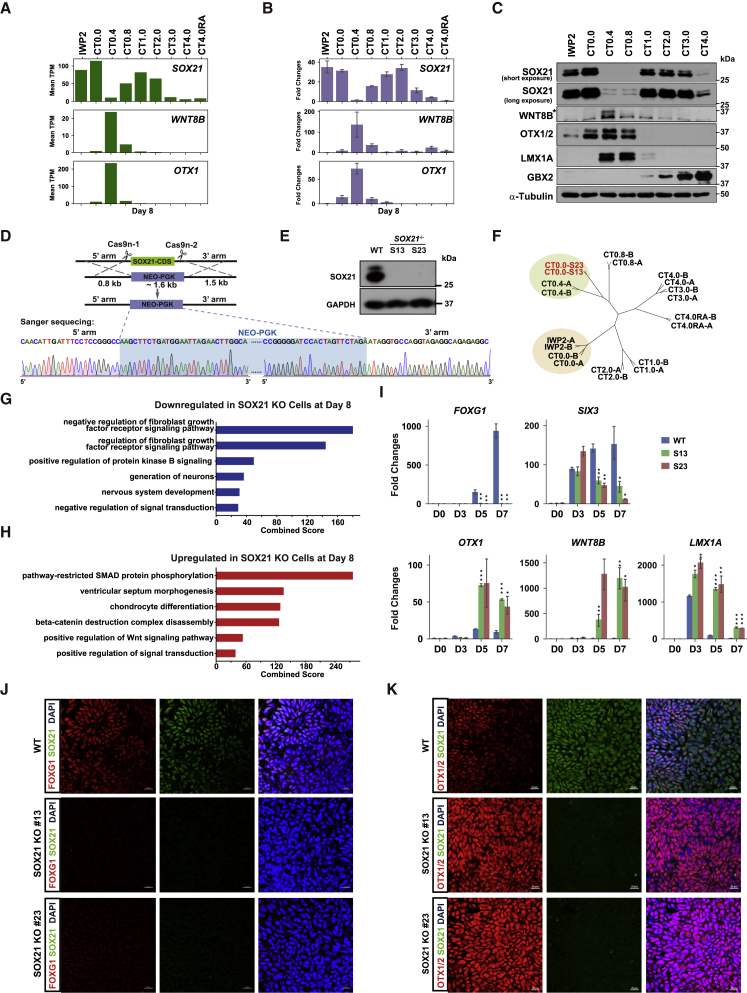
Figure 2.
SOX21 Participates in Forebrain Regionalization
(A) Expression levels of SOX21, WNT8B, and OTX1 at day 8 of rostrocaudal differentiation obtained from our RNA-seq data.
(B) Real-time qPCR validation of SOX21, WNT8B, and OTX1 expression levels at day 8. Data are represented as fold changes relative to undifferentiated WT hESCs (day 0) and shown as means ± SEM; n = 3.
(C) Representative western blot analysis of SOX21, WNT8B, OTX1/2, LMX1A, and GBX2 at day 8. Antibodies of OTX1/2 recognize both OTX1 and OTX2. A star indicates the specific band of WNT8B proteins.
(D) The strategy to establish SOX21 KO clones using the CRISPR/Cas9 nickase with four gRNAs. A map of Sanger sequencing results shows the homology recombination site in SOX21 KO hESCs. The PGK-NEO cassette is highlighted with light blue. CDS, coding sequence.
(E) Representative western blot analysis of SOX21 protein level in hESC-derived NPCs. WT, wild-type cells; S13 and S23 are two SOX21 KO clones.
(F) Hierarchical clustering between SOX21 KO samples (S13 and S23) and WT rostrocaudal differentiation samples at day 8.
(G and H) GO terms enriched for downregulated (G) and upregulated (H) genes in SOX21 KO cells. Only the top 6 GO terms with FDR <0.05 and combined score >15 are listed.
(I) Real-time qPCR validation of DEGs at days 0, 3, 5, and 7 (CT = 0.0 μM). Data are represented as fold changes relative to undifferentiated WT hESCs (day 0) and shown as means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (n = 3).
(J and K) Typical immunofluorescence images of FOXG1, OTX1/OTX2, and SOX21 at day 7. Antibodies against OTX1/OTX2 recognize both OTX1 and OTX2. Scale bars, 20 μm.
See also Figure S2.