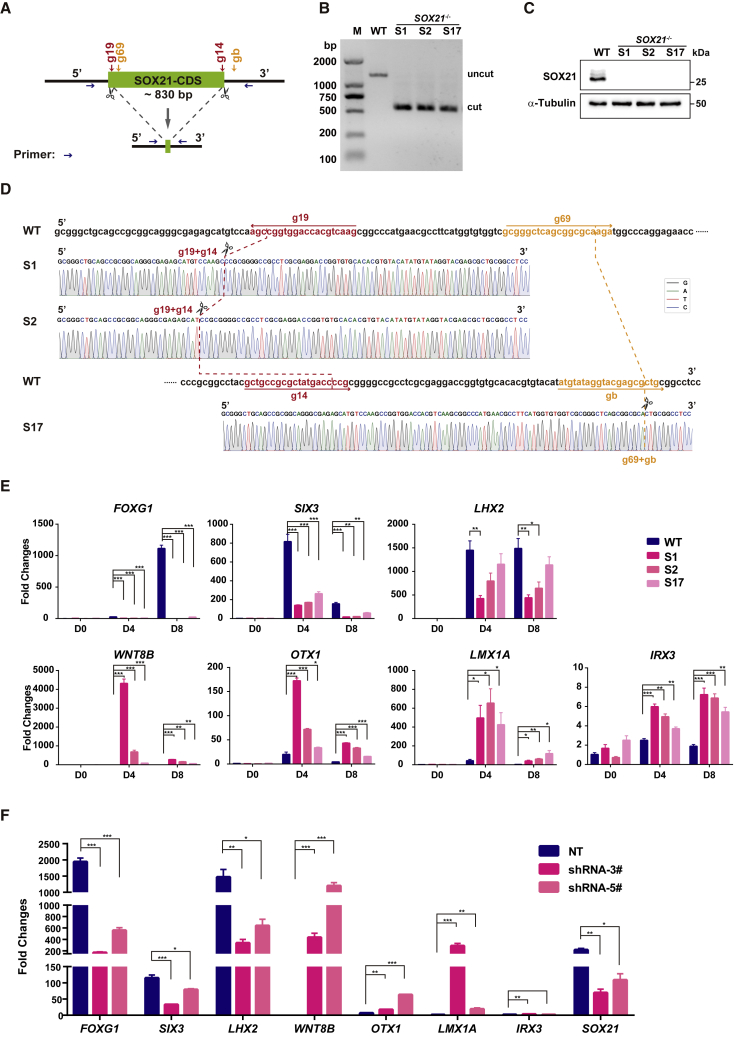
Figure 3.
Generation of Additional SOX21 Knockout hESC Clones
(A) Schematic representation of the paired-KO strategy for SOX21 KO in H9 hESCs. Four different gRNAs targeting different loci were designed (g19 is paired with g14, and g69 is paired with gb). Primers for amplification of genomic DNA are indicated by blue arrows. CDS, coding sequence.
(B) Representative genomic DNA PCR results show the biallelic deletion of SOX21 in hESCs.
(C) Representative western blot analysis of SOX21 protein level in hESC-derived NPCs. WT, wild-type cells; S1, S2 and S17 are three SOX21 KO clones.
(D) Sanger sequencing results show ligation sites (dashed line) in two gRNA pairs (g19 + g14 and g69 + gb).
(E) Results from real-time qPCR analysis of regional marker expression in WT and SOX21−/− NPCs at days 0, 4, and 8. Data are represented as fold changes relative to undifferentiated WT hESCs (day 0) and shown as means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (n = 3).
(F) Results from real-time qPCR analysis of regional marker expression in NT NPCs and SOX21 knockdown NPCs at day 8. For inducible knockdown of SOX21, cells were treated with doxycycline (100 ng/mL) from days 0 to 8. Data are represented as fold changes relative to undifferentiated WT ESCs (day 0) and shown as means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (n = 3). NT, nontarget control.