Figure 4.
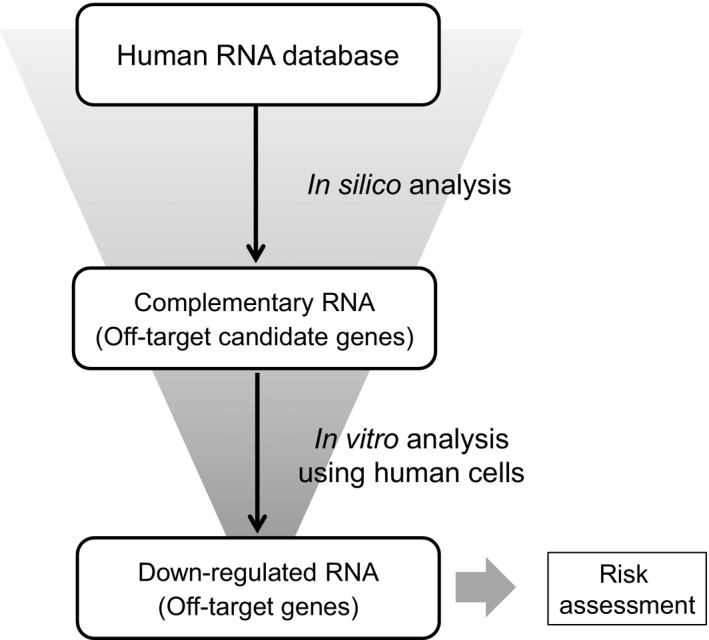
Scheme for the assessment of hybridization‐dependent off‐target effects of gapmer ASOs. In silico analysis: Off‐target candidate genes with complementary RNA sequences are selected from a human RNA database (e.g., D3G) using an appropriate search algorithm (e.g., GGGenome). In vitro analysis using human cells: The ASO is introduced into cultured human cells, and the changes in gene expression of off‐target candidate genes are analyzed. Off‐target candidate genes are narrowed down by considering those with gene expression down‐regulated to <50% as off‐target genes. Risk assessment: The risk of adverse effects emerging from the off‐target genes is investigated by comprehensive consideration of the function, etc., of the gene
