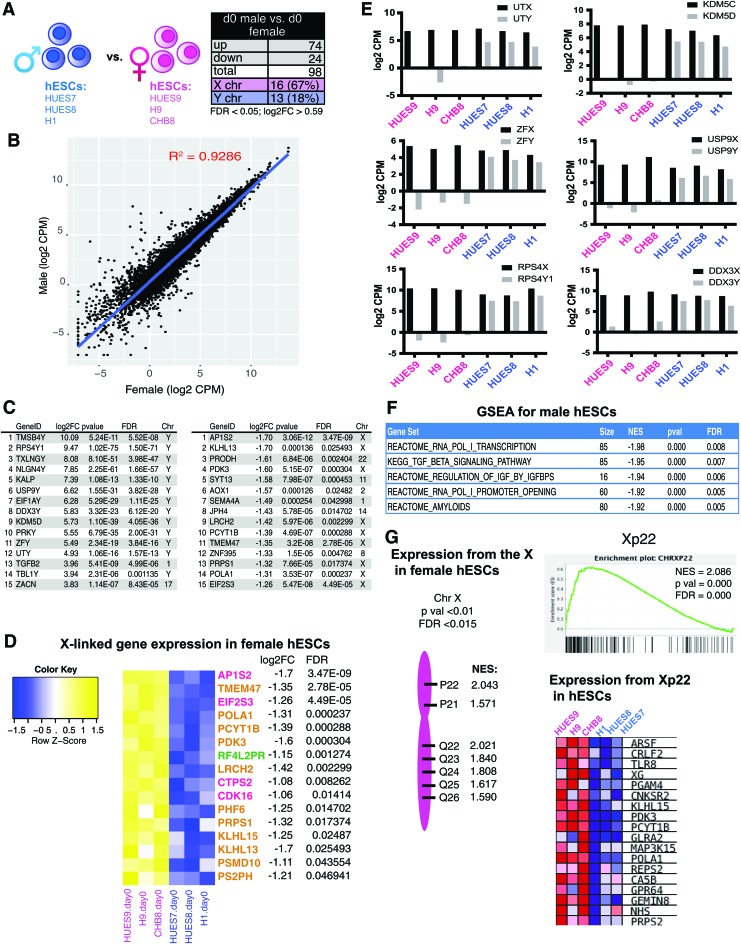
FIG. 2.
Pluripotent hESCs exhibit sex-specific differences with gene expression. (A) Schematic depicting the three male and female hESCs transcriptionally profiled (left) and table showing the total number of significant DEGs between sexes (right; FDR <0.05, log2FC >0.59). (B) Scatterplot of log2 transformed reads (CPMs) of male compared with female hESCs. (C) List of the top 15 significant (P < 0.05, log2FC >0.59) most highly expressed DEGs comparing male (left) with female (right) hESCs. Genes expressed in male hESCs (left); genes expressed in female hESCs (right). (D) Heatmap for X-linked genes overexpressed in female hESCs. Genes in pink denote known XCI escape; green represents variable escape from XCI; yellow represents genes normally subject to XCI. (E) Log2 CPMs for X/Y gene pairs (black bars: X-linked; gray bars: Y-linked) in female (pink) and male (blue) hESCs. (F) GSEA of significantly enriched canonical pathways (C2CP) in male hESCs compared with female hESCs (P < 0.0005). (G) GSEA showing chromosomal locations for X-linked gene sets enriched in female hESCs (P < 0.01, FDR <0.015). CPM, counts per million; DEGs, differentially expressed genes; FDR, false discovery rate; GSEA, Gene Set Enrichment Analysis; XCI, X-chromosome inactivation. Color images available online at www.liebertpub.com/scd