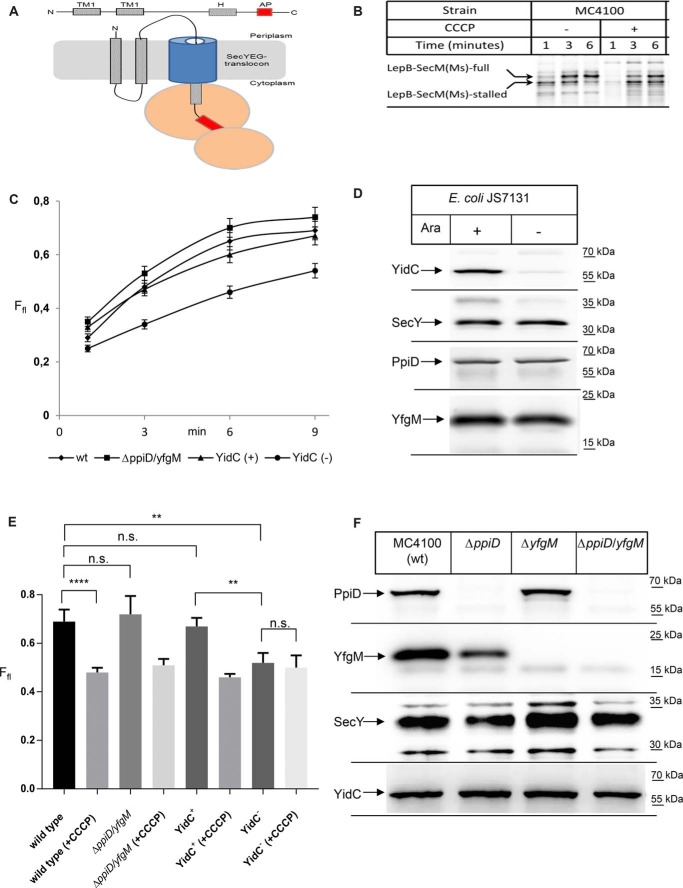
Figure 5.
YidC but not PpiD/YfgM executes a pulling force on nascent SecY substrates. A, cartoon showing the LepB–SecM(Ms) force sensor used in this study. The arrest peptide (AP) is shown in red, and the distance between the arrest peptide and the C terminus is 23 amino acids. B, LepB–SecM(Ms) was expressed in vivo in WT E. coli strain MC4100 and labeled with [35S]methionine/cysteine. After the indicated time, whole cells were precipitated with ice-cold trichloroacetic acid (TCA), and samples were after centrifugation and denaturation in loading dye separated by SDS-PAGE and analyzed by phosphorimaging. Indicated are the full-length LepB–SecM(Ms) and the stalled version, lacking the C-terminal 23 amino acids. When indicated the protonophore CCCP was added for dissipating the proton-motive force. C, as in B, but the ΔppiD/yfgM double deletion strain and the conditional YidC-depletion strain JS7131 were used for expression. JS7131 was grown either in the presence of arabinose (YidC+) or in the presence of glucose (YidC−). The amounts of full-length and stalled LepB were quantified after phosphor-imaging using the ImageQuantTL/ImageJ software, and the fraction of the full-length(Ffl) is displayed. The values correspond to the mean of at least three independent replicates, and the standard deviation is indicated by error bars. D, amounts of YidC, SecY, PpiD, and YfgM in JS7131 cells analyzed in C were determined by Western blotting of TCA-precipitated cells. E, fraction of full-length LepB after 9 min of labeling was analyzed in the indicated strains in the presence or absence of CCCP for dissipating the pmf. Shown are the mean values of at least three independent experiments, and the standard deviations are indicated by the error bars. p values were calculated with an unpaired t test (n ≥3). ****, p value < 0.0001; **, p value < 0.01. n.s. is not significant. F, amounts of PpiD, YfgM, SecY, and YidC in the indicated single and double deletion strains were analyzed as in D.