Figure 1.
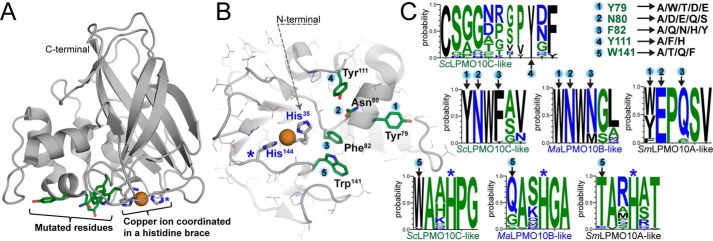
Structural overview of targeted residues in ScLPMO10C. A displays a side view of the crystal structure of the catalytic domain of WT ScLPMO10C (PDB code 4OY7 (34)) with the substrate-binding surface facing downwards. The side chains of residues targeted for mutation are shown with green-colored carbons, and carbons in the side chains of the two histidines that coordinate the copper ion (orange sphere) are colored gray. B shows the substrate-binding surface with residue numbering according to the PDB structure, which is used throughout this study. Note that His-35 is the N-terminal residue of the mature protein and that the mutational work was carried out on the full-length enzyme that includes a family 2 CBM connected by a flexible Pro/Thr-rich linker (13). C shows amino acid frequencies encountered at each mutated position (1–5) in multiple sequence alignments of sequences belonging to various LPMO10 subgroups: ScLPMO10C-like (26 sequences, C1-oxidizing, cellulose); MaLPMO10B-like (28 sequences, C1/C4 oxidation of cellulose and C1 oxidation of chitin); and SmLPMO10A-like (49 sequences, C1-oxidizing, chitin). Position 4 (ScLPMO10C_Y111) is exclusively found in some ScLPMO10C-like sequences and is always present with the upstream motif Asn–Trp–Phe in which the first and the last make up positions 2 and 3 in the library. Mutations included in the library are shown in the top right corner of panel C. Panels A and B were made using PyMOL and the graphs in panel C were generated using WebLogo (36). A blue star in A–C marks the location of the second histidine (His-144) of the histidine brace. See Table S1 for the sequences used in the analyses and Fig. S1 for a structural comparison of natural LPMO10s with experimentally-verified different substrate specificities.