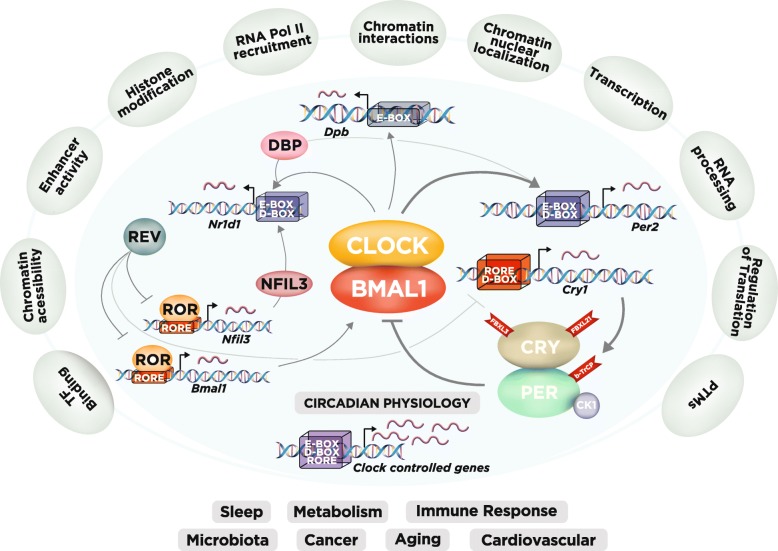
Fig. 2.
The circadian gene network and layers of genome-wide regulation in mammals. At the core of the network, the transcription factors CLOCK and BMAL1 activate the Per1, Per2, Cry1, and Cry2 genes (here we show Per2 and Cry1 as examples), whose protein products (PER and CRY) repress their own transcription. The PER and CRY proteins are post-translationally regulated by parallel E3 ubiquitin ligase pathways (FBXL3 and FBXL21 for CRY and β-TrCP for PER), with PER levels being also regulated by CK1. CLOCK and BMAL1 also regulate the expression of the Nr1d1/2 genes, which encode the nuclear receptors REV-ERBα/β, respectively. These nuclear receptors rhythmically repress the transcription of Bmal1 and Nfil3, two genes that are activated by retinoic acid-related orphan receptor-α/β (RORα/β). In turn, NFIL3 together with D-box binding protein (DBP), as well as CLOCK and BMAL1, regulate a rhythm in the REV-ERBα/β nuclear receptors. These three interlocked transcriptional feedback loops regulate the majority of cycling genes, leading to rhythms in various different physiological systems, from sleep to metabolism and aging (bottom of figure). Note that the E- and D-boxes and the RORE-binding regions are in cis upstream at the promoter; however, they are represented here as a stacked box for simplicity. Recent work has identified additional levels of regulation of circadian gene expression (outer layer of regulation in the figure), including rhythmic histone modifications, RNA polymerase II (Pol II) recruitment, circadian chromosomal conformation interactions and post-translational modifications (PTMs). Please refer to Table S1 for many of the studies that allowed the external regulatory layers to be added to the comprehensive view of the clock