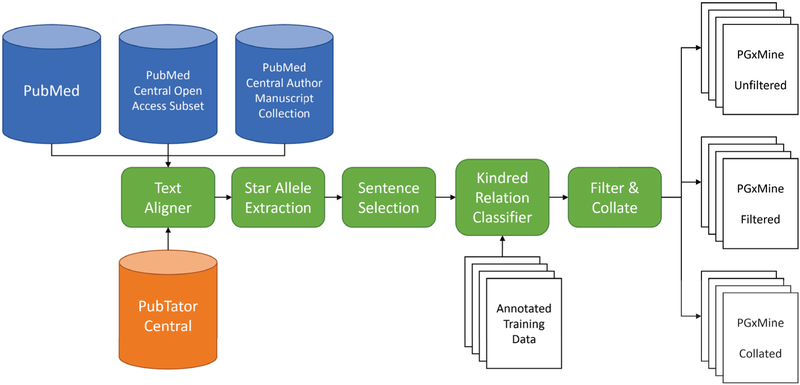
Fig. 1.
An overview of the full PGxMine system. The input data sources on the left (PubMed, etc) and PubTator Central are combined through a text alignment process to identify mentions of specific biological entities in published literature. Star Alleles (e.g. CYP2D6*2) are then found using gene annotations. Sentences are filtered using keywords to enrich for pharmacogenomic topics. A Kindred supervised classifier is then trained and applied to identify specific variant/chemical associations. These are then filtered for high probability matches and collated to produce the three output files on the right.