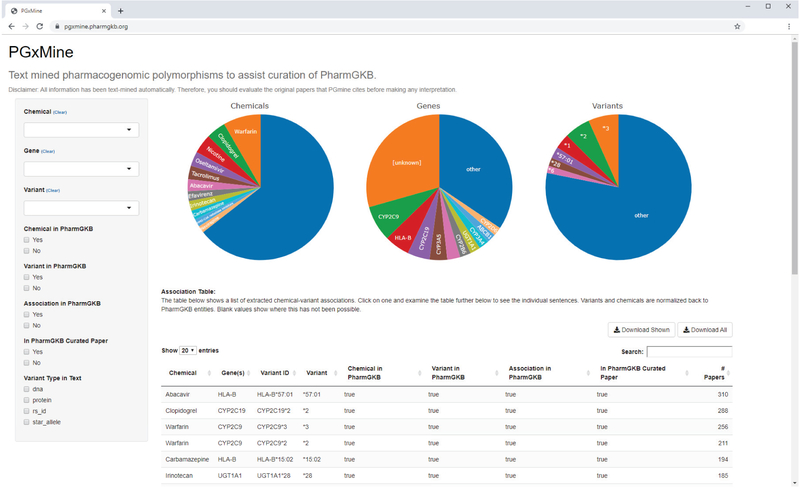
Fig. 2.
The data can be viewed through an R Shiny application in a web browser which allows the user to sort and filter the data and explore specific sentences that mention the association. The main table shows chemical/variant associations with gene information where possible. This table can be filtered using controls to the left to select specific chemicals, variants or genes. It can also be filtered for whether elements are already curated into PharmGKB. By selecting a row in the top table, a further table below (not shown) is populated with sentences that mention the selected association along with paper metadata.