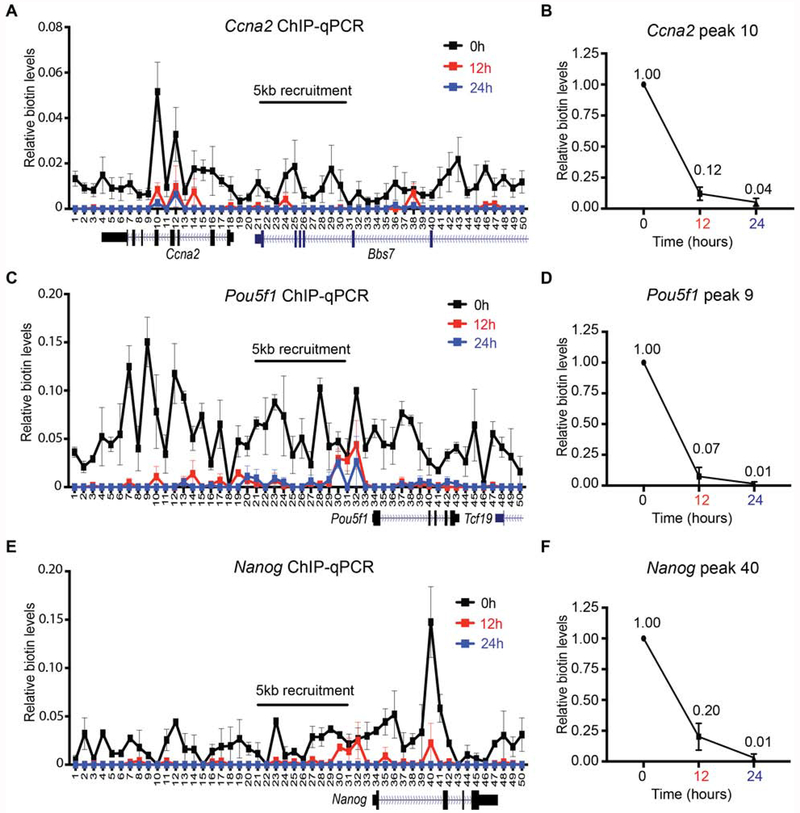
Figure 3. Dispersed segregation of active parental H3 domains.
Native mononucleosomal biotin ChIP-qPCR from G1/S-blocked mESCs that were released following a 6 hr Dox and exogenous biotin pulse, with BirA targeted to the Ccna2 (A, B), Pou5f1 (C, D), and Nanog (E, F) loci. Data shown is the average of 3 biological replicates spanning a 25 kb area at a resolution of 500 bp. For (B) (D), and (F), the graph highlights the biotin enrichment taken from the highest peak of the corresponding assays: Ccna2 primer set 10 (A), Pou5f1 primer set 9 (C), and Nanog primer set 40 (E). All biotin enrichment levels are relative to input, normalized to Drosophila chromatin spike-in, followed by subtraction of the -Dox control. Error bars represent standard error. For panels (B), (D), and (F), the dataset for 0 hr was set to 1.