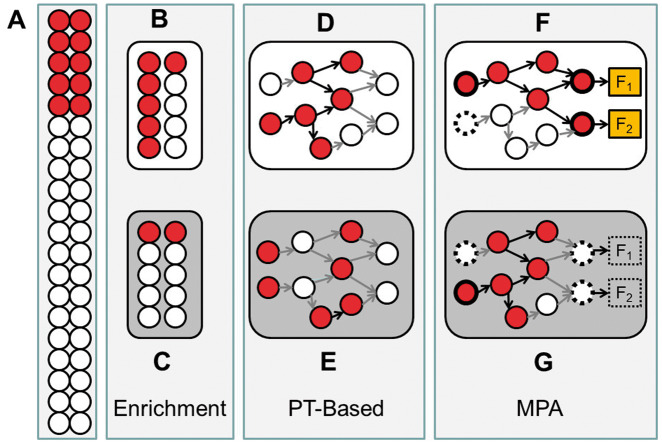
Figure 1.
Schematic representation of the three families of methods: enrichment analysis, PT-based analysis and MPA. The conventional enrichment analysis assumes the existence of a background (A) in which an observed percentage (25% in the example) of the genes differentially expressed (or mutated, associated to a trait, etc.). If gene sets are sampled based on some property shared by all the genes (e.g. they belong to a given pathway), a scenario (B) in which 60% of them are differentially expressed is found; the application of a simple test will evidence that this gene set is significantly enriched in differentially expressed genes, whereas in other scenarios (C), the gene set would not be different from a random sample of genes from the background. A PT-based algorithm takes into consideration the topology of the gene set, and a scenario (C) in which the differentially expressed genes are more connected among them would get a better score that an alternative scenario (D) in which the level of connection of the genes is lower. The significance of this data set would depend on the algorithm that estimates the score and the specific test applied. In MPA, there is more or less specific definition of circuits (subnetworks) within the pathway that should be related to cell activity in some way, and the connectivity of such circuits will determine the potential changes in cell activity. If circuits define subnetworks connecting receptor proteins to effector proteins in a signal transduction pathway, the same number of active genes could allow signal transduction (E) or being incompatible with the arrival of the signal to the current effector proteins (F), even in scenarios that would be significantly enriched in a conventional enrichment method.