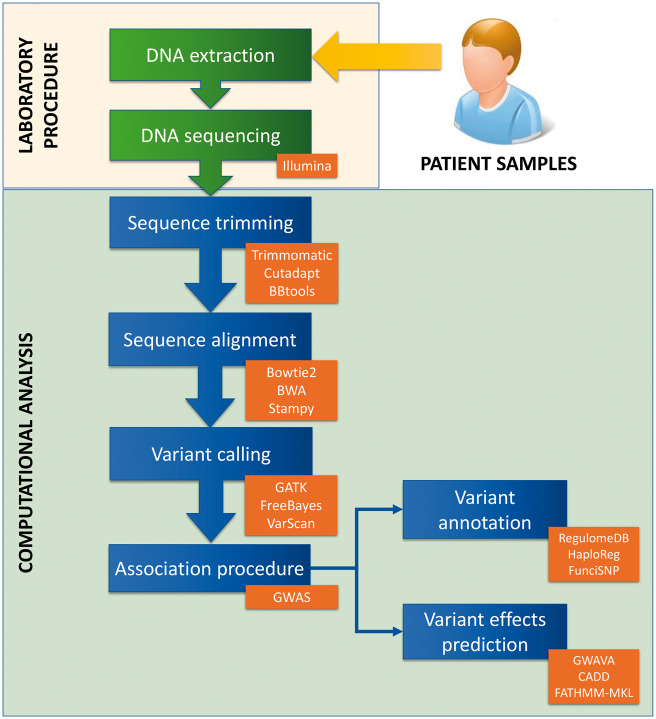
Figure 1.
Description of the steps necessary to characterize regulatory variants. Following DNA extraction and sequencing of individual samples, a number of computational steps are performed. First, trimming and alignment of the sequences is necessary. Then a file with all the variants is obtained using the variant calling procedure. At this point, these variants must be associated with disease, using GWAS or other experimental designs such as trio analysis. Next the non-coding SNPs can be analysed and annotated putatively based on overlap with genomic functional elements (variant annotation) and their putative effects can be predicted (variant effects prediction). In orange boxes, there are some examples of tools or analysis types that can be used for each step, and in the case of variant annotation and variant effects prediction, we show examples of tools focused on regulatory variants.