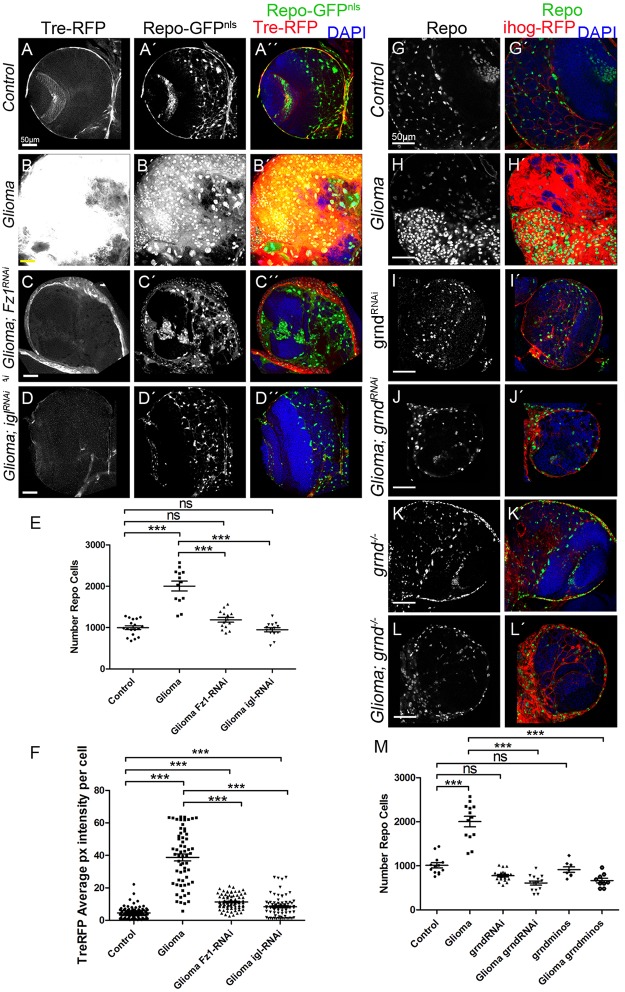
Fig 6. JNK signaling pathway is activated in glioma via the TNF receptor Grindelwald which mediates GB progression.
Glial cell nuclei are labeled with UAS-GFPNLS (gray or green in the merge) driven by repo-Gal4. (A–D) JNK signaling pathway reporter TRE-RFP (gray or red in the merge) in control, glioma, glioma Gap43-RNAi, and Glioma Fz1-RNAi brain sections. The number of glial cells is quantified in panel E, and the TRE-RFP average pixel intensity per glial cells is quantified in panel F. (G–L) Glia are labeled with UAS-Ihog-RFP (red) driven by repo-Gal4 to visualize active cytonemes/TM structures in glial cells and stained with Repo (gray or green in the merge) in the following genotypes: control, glioma, grnd-RNAi, glioma grnd-RNAi, grnd−/− and Glioma grnd−/− brain sections. The number of Repo+ cells is quantified in panel M. Nuclei are marked with DAPI. Error bars show SD; *P < 0.01, **P < 0.001, ***P < 0.0001, or ns for nonsignificant. Scale bar size is indicated in this and all figures. The data underlying this figure can be found in S1 Data. Genotypes: (A) TRE-RFP; repo-Gal4, UAS-GFPNLS/UAS-lacZ, (B) UAS-dEGFRλ, UAS-dp110CAAX; TRE-RFP; repo-Gal4, UAS-GFPNLS, (C) UAS-dEGFRλ, UAS-dp110CAAX; TRE-RFP; repo-Gal4, UAS-GFPNLS/UAS-Gap43-RNAi, (D) UAS-dEGFRλ, UAS-dp110CAAX; TRE-RFP/UAS-Fz1-RNAi; repo-Gal4, UAS-GFPNLS, (G) repo-Gal4, ihog-RFP/UAS-lacZ, (H) UAS-dEGFRλ, UAS-dp110CAAX;; repo-Gal4, UAS-ihog-RFP, (I) UAS-grnd-RNAi; repo-Gal4, ihog-RFP, (J) UAS-dEGFRλ, UAS-dp110CAAX; UAS-grnd-RNAi; repo-Gal4, UAS-ihog-RFP, (K) grndMINOS/grndMINOS; repo-Gal4, ihog-RFP, (L) UAS-dEGFRλ, UAS-dp110CAAX; grndMINOS/grndMINOS; repo-Gal4, UAS-ihog-RFP. Fz1, Frizzled1; GB, glioblastoma; GFP, green fluorescent protein; grnd, Grindelwald; Ihog, interference hedgehog; JNK, cJun N-terminal kinase; TM, tumor microtube; TRE-RFP, RFP fluorescent protein regulated by a transcriptional response element of JNK pathway.