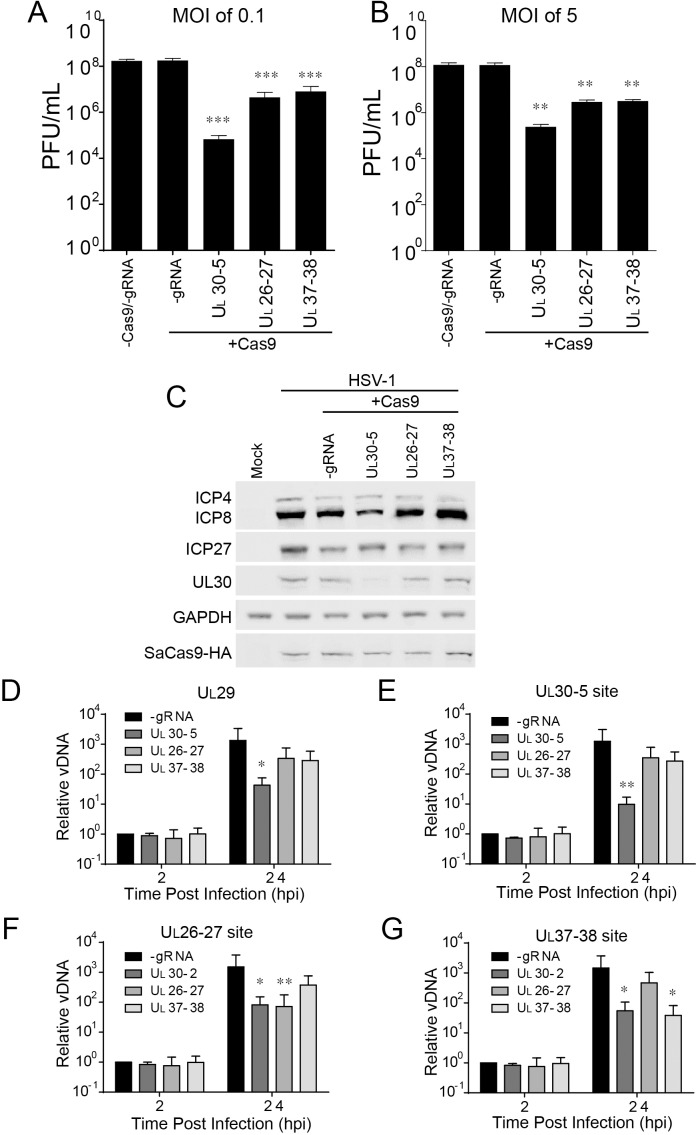
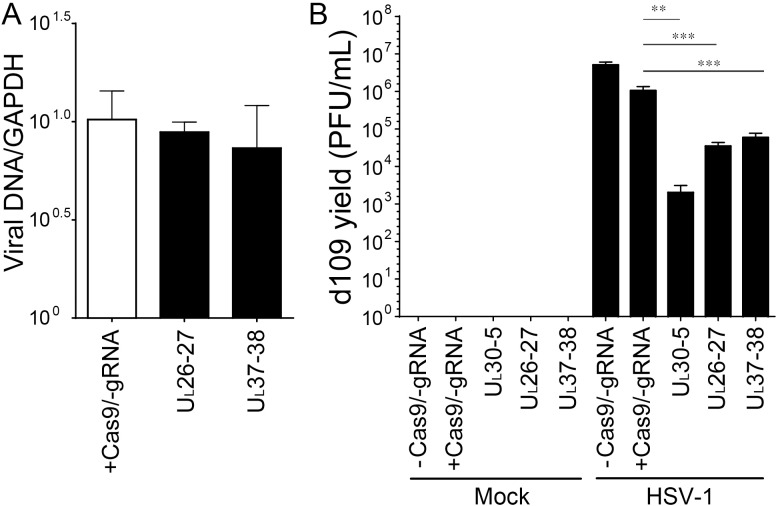
Figure 8. Effect of CRISPR-Cas9 on non-coding and non-essential regions of the HSV-1 genome.
(A and B) HFFs transduced with lentivirus expressing SaCas9 and sgRNAs were infected with HSV-1 at an MOI of 0.1 (A) or 5 (B), harvested at 48 hpi or 24 hpi respectively. Viral yields were determined by plaque assays. The histogram shows the mean values and standard deviations from biological replicates (N = 4). (C) HFFs transduced with lentivirus expressing SaCas9 and sgRNA were infected with HSV-1 at an MOI of 5 and harvested at 10 hpi. Proteins were detected using immunoblotting with antibodies specific for the indicated proteins. GAPDH was shown as a control. (D–G) HFFs transduced with lentivirus expressing SaCas9 and sgRNA were infected with HSV-1 at an MOI of 3 and harvested at the indicated time post infection. The accumulated DNAs were detected by real time PCR amplifying in the UL29 gene (ICP8, (D) or over the UL30-5 (E), UL26-27 (F), and UL37-38 (G) sgRNA target sites. The histogram shows the mean values and standard deviations from biological replicates (N = 4, (A and B) one-way ANOVA with Dunnett’s multiple comparisons test, (D–G) Ratio paired t test, *p<0.05, **p<0.01, and ***p<0.001).